
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,792,823 – 4,792,989 |
| Length | 166 |
| Max. P | 0.999032 |

| Location | 4,792,823 – 4,792,925 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 98.69 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4792823 102 + 27905053 CAACCACAACCGAACAAACAACAAAAAACAAACAACAGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAAAUGCCGUG ...........................................(((...((((((......((((((((.....)))))))).....))))))..))).... ( -19.50) >DroSec_CAF1 28244 102 + 1 CAACCACAACCGAACAAACAACACAAAACAAACAACAGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUG ...........................................(((...((((((......((((((((.....)))))))).....))))))..))).... ( -20.60) >DroSim_CAF1 23651 102 + 1 CAACCACAACCGAACAAACAACACAAAACAAACAACAGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUG ...........................................(((...((((((......((((((((.....)))))))).....))))))..))).... ( -20.60) >consensus CAACCACAACCGAACAAACAACACAAAACAAACAACAGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUG ...........................................(((...((((((......((((((((.....)))))))).....))))))..))).... (-20.23 = -20.23 + -0.00)



| Location | 4,792,823 – 4,792,925 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 98.69 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4792823 102 - 27905053 CACGGCAUUUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCUGUUGUUUGUUUUUUGUUGUUUGUUCGGUUGUGGUUG ((((((....((((((.(..((((((((((..((((.(((.....))).))))..)))))))...)))..)........)))))).......)))))).... ( -28.50) >DroSec_CAF1 28244 102 - 1 CACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCUGUUGUUUGUUUUGUGUUGUUUGUUCGGUUGUGGUUG ((((((....((((((((..((((((((((..((((.(((.....))).))))..))))))..........))))..)))))))).......)))))).... ( -30.10) >DroSim_CAF1 23651 102 - 1 CACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCUGUUGUUUGUUUUGUGUUGUUUGUUCGGUUGUGGUUG ((((((....((((((((..((((((((((..((((.(((.....))).))))..))))))..........))))..)))))))).......)))))).... ( -30.10) >consensus CACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCUGUUGUUUGUUUUGUGUUGUUUGUUCGGUUGUGGUUG ((((((....((((((((..((((((((((..((((.(((.....))).))))..))))))..........))))..)))))))).......)))))).... (-29.40 = -29.73 + 0.33)



| Location | 4,792,859 – 4,792,949 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -26.14 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4792859 90 + 27905053 AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAAAUGCCGUGUGCUGUUCCAUGCUGUGUGUGCCA .......(((...((((((......((((((((.....)))))))).....)))))).((((.(((((......))))).)))).))).. ( -24.40) >DroSec_CAF1 28280 90 + 1 AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUGUGCUGUUCCAUACUGUGUGUGCCA .((((.(((.....)))........((((((((.....))))))))....))))(((.((((.(((((......))))).)))).))).. ( -27.20) >DroSim_CAF1 23687 90 + 1 AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUGUGCUGUUCCAUGCUGUGUGUGCCA .((((.(((.....)))........((((((((.....))))))))....))))(((.((((.(((((......))))).)))).))).. ( -26.20) >DroEre_CAF1 28567 90 + 1 AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUGUGCUGUUCCAUGCUGUGUGUGCCA .((((.(((.....)))........((((((((.....))))))))....))))(((.((((.(((((......))))).)))).))).. ( -26.20) >DroYak_CAF1 28921 90 + 1 AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUGUGCUGCGCCAUGCCGUGUGUGCCA (((((..(((...((((((......((((((((.....)))))))).....))))))..)))..))).))(((((((...))).)))).. ( -28.10) >consensus AGACAAUGCAAAAUGCAAUUGAGAUGCAAAUGCAAAUUGCAUUUGCCACAUGUUGCAGAUGCCGUGUGCUGUUCCAUGCUGUGUGUGCCA .((((.(((.....)))........((((((((.....))))))))....))))(((.((((.(((((......))))).)))).))).. (-26.14 = -25.98 + -0.16)



| Location | 4,792,859 – 4,792,949 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4792859 90 - 27905053 UGGCACACACAGCAUGGAACAGCACACGGCAUUUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .((((......((........)).....(((..(((((..((..((((((((.....))))))))....)).)))))...)))..)))). ( -24.90) >DroSec_CAF1 28280 90 - 1 UGGCACACACAGUAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .((((......((.((......)).)).(((..(((((..((..((((((((.....))))))))....)).)))))...)))..)))). ( -23.50) >DroSim_CAF1 23687 90 - 1 UGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .((((......((........)).....(((..(((((..((..((((((((.....))))))))....)).)))))...)))..)))). ( -24.20) >DroEre_CAF1 28567 90 - 1 UGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .((((......((........)).....(((..(((((..((..((((((((.....))))))))....)).)))))...)))..)))). ( -24.20) >DroYak_CAF1 28921 90 - 1 UGGCACACACGGCAUGGCGCAGCACACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .(((.....(.(((((..((((..........))))..))))).)..(((((((..((((.(((.....))).))))..)))))))))). ( -28.20) >consensus UGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAUGCAAUUUGCAUUUGCAUCUCAAUUGCAUUUUGCAUUGUCU .((((......((........)).....(((..(((((..((..((((((((.....))))))))....)).)))))...)))..)))). (-24.08 = -23.92 + -0.16)

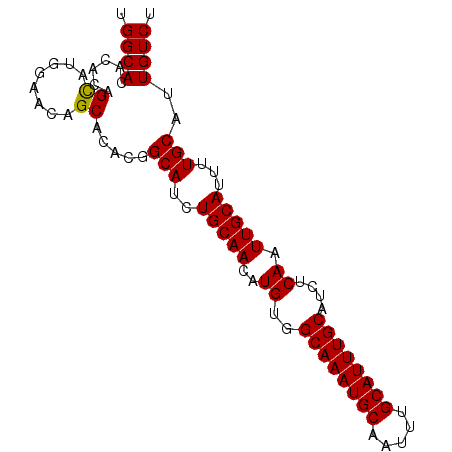

| Location | 4,792,899 – 4,792,989 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.76 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.20 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4792899 90 - 27905053 UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACAGCAUGGAACAGCACACGGCAUUUGCAACAUGUGGCAAAU ((((((((((((((........((((......)))).(((((.......))))).......)))))).((....))..))).)))))... ( -23.30) >DroSec_CAF1 28320 90 - 1 UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACAGUAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAU ((((((((((((((........((((......)))).(((((.......))))).......)))))).((....))..))).)))))... ( -21.30) >DroSim_CAF1 23727 90 - 1 UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAU ((((((((((((((........((((......)))).(((((.......))))).......)))))).((....))..))).)))))... ( -23.30) >DroEre_CAF1 28607 90 - 1 UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAU ((((((((((((((........((((......)))).(((((.......))))).......)))))).((....))..))).)))))... ( -23.30) >DroYak_CAF1 28961 90 - 1 UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACGGCAUGGCGCAGCACACGGCAUCUGCAACAUGUGGCAAAU ........((((((........((((......))))......)))))).(.(((((..((((..........))))..))))).)..... ( -22.74) >consensus UGUUAAUGGUGUGCAUUAUUUUCAUGUUUAUUUAUGUUGCUGGCACACACAGCAUGGAACAGCACACGGCAUCUGCAACAUGUGGCAAAU ((((((((((((((........((((......)))).(((((.......))))).......)))))).((....))..))).)))))... (-23.08 = -22.76 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:17 2006