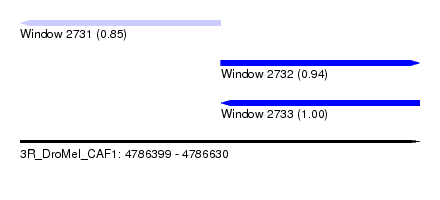
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,786,399 – 4,786,630 |
| Length | 231 |
| Max. P | 0.999140 |

| Location | 4,786,399 – 4,786,515 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -17.70 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4786399 116 - 27905053 -AAAAAUUAAAAAACUCCAGCAACAAUGGCAACAACAACAGUAGCGGCUGUUUGUACCGUUAGAUGACGCAAAAUUACGCUUACUCAUUCGGCAAGCGGCAGUUCCCUC--AAAAACAA -...........((((((.((.....((....))((((((((....)))).))))...((..(((((.((........))....)))))..))..)))).)))).....--........ ( -20.90) >DroEre_CAF1 21915 115 - 1 --AAAAUUUUAAAACACCAGCAACAAUGGCAACAACAACAGUAGCGGCCGUAUGUACCGUUAGAUGACGCAAAAUUACGCUUACUCAUUCGGCAAGCGGCAGUUCCCCC--AAAAACAA --..............((.((.((((((((..(.((....)).)..))))).)))...((..(((((.((........))....)))))..))..))))..........--........ ( -19.80) >DroAna_CAF1 21237 93 - 1 G--------------------------AACAACAACAACAGUAGCGGCUGCUUGUACCGUUAGAUGACGCAAAAUUACGCUUACUCAUUCGGCAAGCAGCGUUCCCCCCCAAAAAACAA (--------------------------(((....((....))....((((((((..(((...(((((.((........))....))))))))))))))))))))............... ( -21.50) >consensus __AAAAUU__AAAAC_CCAGCAACAAUGGCAACAACAACAGUAGCGGCUGUUUGUACCGUUAGAUGACGCAAAAUUACGCUUACUCAUUCGGCAAGCGGCAGUUCCCCC__AAAAACAA ..............................................((((((((..(((...(((((.((........))....))))))))))))))))................... (-17.70 = -17.37 + -0.33)

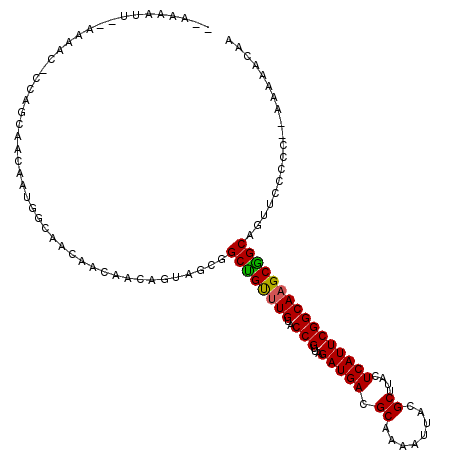

| Location | 4,786,515 – 4,786,630 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4786515 115 + 27905053 -----UUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAGCCGAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU -----....((((((((....((......)).....))))))))..(((((((......(((((...(((((.......))))).))))).((((.(((....))).)))).))))))). ( -25.80) >DroSec_CAF1 21773 116 + 1 ----UUUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAACCGAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU ----......((((.......))))..(((((......(((((.((((((((........(((.......))).))).))))).)))))..((((.(((....))).))))..))))).. ( -22.00) >DroSim_CAF1 17237 119 + 1 UUUUUUUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAGCCGAU-GUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU .........((((((((....((......)).....)))))))).-(((((((......(((((...(((((.......))))).))))).((((.(((....))).)))).))))))). ( -25.80) >DroEre_CAF1 22030 115 + 1 -----UUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAGCCGAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU -----....((((((((....((......)).....))))))))..(((((((......(((((...(((((.......))))).))))).((((.(((....))).)))).))))))). ( -25.80) >DroYak_CAF1 22387 118 + 1 --UUUUUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAGCCUAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU --.........((((((....((......)).....))))))....(((((((......(((((...(((((.......))))).))))).((((.(((....))).)))).))))))). ( -24.00) >DroAna_CAF1 21330 119 + 1 UUUUUUUUUUUGGCCGCCAUUGCCAUUCCGUUCUUAC-GGCCGAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAGCAAACGGUUUUAUGAUUUAUUUUUAGCUCGUCCAACGGCU .........(((((((.....((......)).....)-))))))..(((((((.......(((.......))).(((.(((.((..((..........))..)).))).)))))))))). ( -24.20) >consensus ____UUUUUUUGGCUGGCAUUGCCAUUCCGUUCUUACCAGCCGAUUGUUGUUGUAUUUUAGACCAUUUUUGUCAGACAAACAAACGGUUUUAUGGUUUAUUUUUAGCCCGUCCAACGGCU .........((((((((....((......)).....))))))))..(((((((......(((((...(((((.......))))).))))).((((.(((....))).)))).))))))). (-23.41 = -23.38 + -0.02)
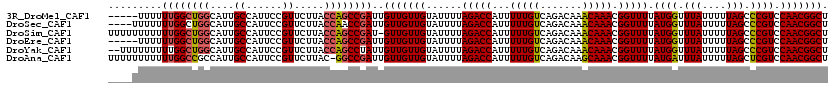
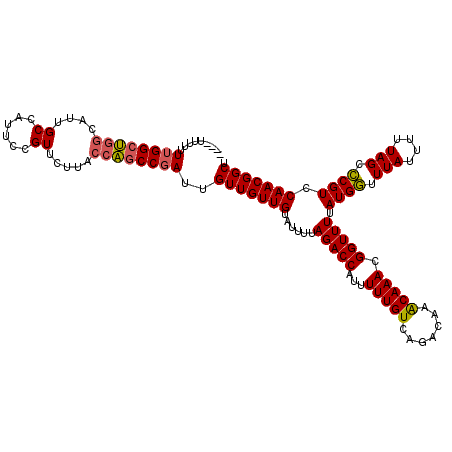
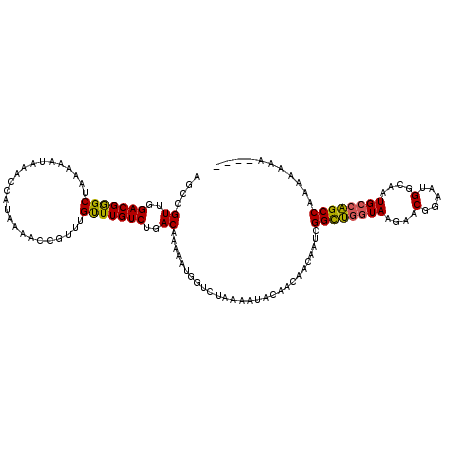
| Location | 4,786,515 – 4,786,630 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.08 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4786515 115 - 27905053 AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUCGGCUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAA----- .((((((.(.(((((...((((((((.........))))))))))))).)...))))))...................((((((((....(.....)....))))))))......----- ( -27.20) >DroSec_CAF1 21773 116 - 1 AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUCGGUUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAAA---- .((((((.(.(((((...((((((((.........))))))))))))).)...))))))...................((((((((....(.....)....)))))))).......---- ( -24.80) >DroSim_CAF1 17237 119 - 1 AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAAC-AUCGGCUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAAAAAAA .((((((.(.(((((...((((((((.........))))))))))))).)...))))))...............-...((((((((....(.....)....))))))))........... ( -27.20) >DroEre_CAF1 22030 115 - 1 AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUCGGCUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAA----- .((((((.(.(((((...((((((((.........))))))))))))).)...))))))...................((((((((....(.....)....))))))))......----- ( -27.20) >DroYak_CAF1 22387 118 - 1 AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUAGGCUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAAAAA-- .((((((.(.(((((...((((((((.........))))))))))))).)...))))))...................((((((((....(.....)....)))))))).........-- ( -27.10) >DroAna_CAF1 21330 119 - 1 AGCCGUUGGACGAGCUAAAAAUAAAUCAUAAAACCGUUUGCUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUCGGCC-GUAAGAACGGAAUGGCAAUGGCGGCCAAAAAAAAAAA ....((..(((((((........................)))))))..)).....(((((....................((-((....))))....((....))))))).......... ( -26.96) >consensus AGCCGUUGGACGGGCUAAAAAUAAACCAUAAAACCGUUUGUUUGUCUGACAAAAAUGGUCUAAAAUACAACAACAAUCGGCUGGUAAGAACGGAAUGGCAAUGCCAGCCAAAAAAA____ ....((..(((((((........................)))))))..))............................((((((((....(.....)....))))))))........... (-24.43 = -24.08 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:04 2006