
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,784,825 – 4,785,077 |
| Length | 252 |
| Max. P | 0.998324 |

| Location | 4,784,825 – 4,784,937 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.69 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526182 |
| Prediction | RNA |
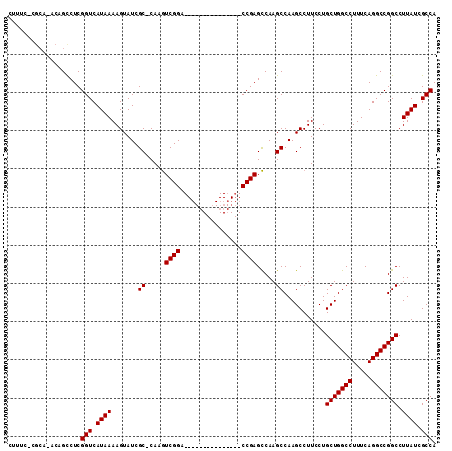
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4784825 112 - 27905053 CUUUCCCGCAAACAGUCUCGGUCAUAAAAGUAUCGCCCAAAUCGGGCCAGGCUAUGGCGAGCCGAGUCAAGCCAAGCCUUCCUGCUGGCCUUUCAGGCCGGCCUUAUCGCCA ...................(((.((((.......((((.....)))).(((((.((((............)))))))))....(((((((.....))))))).)))).))). ( -37.80) >DroSec_CAF1 20047 84 - 1 ------------CAGCCUCGGUCAUAAAAGUAUCGC-CAAGUCGGA---------------CCGAGCCAAGCCAAGCCUUCCUGCUGGCCUUUCAGGCCGGCCUUAUCGCCA ------------..(.(((((((......(.....)-.......))---------------))))).).......((......(((((((.....)))))))......)).. ( -25.42) >DroSim_CAF1 15509 96 - 1 CUUUCUCGCAUACAGCCUCGGUCAUAAAAGUAUCGC-CAAGUCGGA---------------CCGAGCCAAGCCAAGCCUUCCUGCUGGCCUUUCAGGCCGGCCUUAUCGCCA .......((.....(.(((((((......(.....)-.......))---------------))))).)..(((.(((......)))((((.....)))))))......)).. ( -26.92) >consensus CUUUC_CGCA_ACAGCCUCGGUCAUAAAAGUAUCGC_CAAGUCGGA_______________CCGAGCCAAGCCAAGCCUUCCUGCUGGCCUUUCAGGCCGGCCUUAUCGCCA ...................(((.((((.......((.....((((................)))).....))...........(((((((.....))))))).)))).))). (-22.69 = -22.69 + -0.00)


| Location | 4,784,937 – 4,785,037 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -33.58 |
| Energy contribution | -33.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4784937 100 - 27905053 CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGUCCACU--ACCCGUGCAGAGUGCCA--GUUCCAGUACAUCU ......(((((((....((.(.((((((((((.(.(((((((....))))))).).)))).))))))).--))..)))))))((((..--......)))).... ( -38.80) >DroSec_CAF1 20131 97 - 1 CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGUCCACU--ACCCAUGAAGAGUGCCAGAGUUCCAGUA----- ...............(((..(((.(((((((((((((((((((....(((((...))))).))))))))--.))).))..))).)))..)))..)))..----- ( -33.80) >DroSim_CAF1 15605 102 - 1 CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGUCCACU--ACCCAUGAAGAGUGCCAGAGUUCCAGUACAUCU .............((.(((.((((((((((((.(.(((((((....))))))).).))))))...((((--..........)))).......))))))))).)) ( -35.00) >DroEre_CAF1 20409 99 - 1 CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUUCGCUGCGUGGAGGGUCCACUG---CCAUGCAGAGUGCCA--GUUCCAGUACAUCU ......(((((((.((....(.((((((((((.(.(((((((....))))))).).)))).))))))).)---).)))))))((((..--......)))).... ( -37.00) >DroYak_CAF1 20742 102 - 1 CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGUCCACUCUACCAAUGCAGAGUGCCA--GUUCCAGUACAUCU ...............(((..(((((.((((((.(.(((((((....))))))).).))))))...((((((........)))))))))--))..)))....... ( -40.60) >consensus CAAAAAUUUGCAUAGUUGUUGCUGGGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGUCCACU__ACCCAUGCAGAGUGCCA__GUUCCAGUACAUCU ......((((((((((....)))(((((((((.(.(((((((....))))))).).)))).))))).........)))))))((((..........)))).... (-33.58 = -33.86 + 0.28)



| Location | 4,784,973 – 4,785,077 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.77 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -19.78 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4784973 104 + 27905053 ACCCUCCACGCAGCGGAUCCAAAUCCACUCCCUGGAGGCC----------------CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGAUGCUUAACAAUUUUUAGUCGCCGCUU ..((((((.(.((.((((....)))).)).).))))))..----------------.((((.(((((((((((....))))))))).....)).))))...................... ( -26.00) >DroSec_CAF1 20164 104 + 1 ACCCUCCACGCAGCGGAUCCAAAUCCACUCCCUGGAGGCC----------------CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGAUGCUUAACUAUUUUUAGUCGCCGCUU ..((((((.(.((.((((....)))).)).).))))))..----------------.(((...((((((((((....))))))))))........((...((((....)))).)).))). ( -28.00) >DroSim_CAF1 15643 104 + 1 ACCCUCCACGCAGCGGAUCCAAAUCCACUCCCUGGAGGCC----------------CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGAUGCUUAACUAUUUUUAGUCGCCGCUU ..((((((.(.((.((((....)))).)).).))))))..----------------.(((...((((((((((....))))))))))........((...((((....)))).)).))). ( -28.00) >DroEre_CAF1 20444 104 + 1 ACCCUCCACGCAGCGAAUCCAAAUCCACUCCCUGGAGGCC----------------CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGAUGCUUAACUAUUUUUAGUCGCUGCUU .........(((((((...........(((....)))...----------------.((((.(((((((((((....))))))))).....)).)))).............))))))).. ( -27.20) >DroYak_CAF1 20780 104 + 1 ACCCUCCACGCAGCGGAUCCAAAUCCACUCCCUGGAGGCC----------------CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGACGCUUAACUAUUUUUAGUCGCCGCUC ..((((((.(.((.((((....)))).)).).))))))..----------------.(((...((((((((((....))))))))))........((...((((....)))).)).))). ( -28.30) >DroAna_CAF1 20095 110 + 1 ACCCUCCACGCUG---------AUACACUCCCUAGAGGCCGCUGCCUAGUGGACCCAGGCAACAACUAUGCAAAUUUGUGUAUUUUUUUUAUGAUGCUUAACUAUUUUUAGUCAGU-UUU .........((((---------((..........(.(((((((....))))).)))(((((.((...(((((......)))))........)).)))))...........))))))-... ( -20.00) >consensus ACCCUCCACGCAGCGGAUCCAAAUCCACUCCCUGGAGGCC________________CAGCAACAACUAUGCAAAUUUUUGUGUAGUUUUUAUGAUGCUUAACUAUUUUUAGUCGCCGCUU ..((((((.(.((.((((....)))).)).).))))))...................((((.(((((((((((....))))))))).....)).))))..((((....))))........ (-19.78 = -21.50 + 1.72)



| Location | 4,784,973 – 4,785,077 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.77 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -29.93 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4784973 104 - 27905053 AAGCGGCGACUAAAAAUUGUUAAGCAUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG----------------GGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGU .(((((((((((.(((((..........................)))))...))))))))))).----------------..((((((.(.(((((((....))))))).).)))))).. ( -37.77) >DroSec_CAF1 20164 104 - 1 AAGCGGCGACUAAAAAUAGUUAAGCAUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG----------------GGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGU .(((((((((((....(((((............)))))..(((....)))..))))))))))).----------------..((((((.(.(((((((....))))))).).)))))).. ( -38.30) >DroSim_CAF1 15643 104 - 1 AAGCGGCGACUAAAAAUAGUUAAGCAUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG----------------GGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGU .(((((((((((....(((((............)))))..(((....)))..))))))))))).----------------..((((((.(.(((((((....))))))).).)))))).. ( -38.30) >DroEre_CAF1 20444 104 - 1 AAGCAGCGACUAAAAAUAGUUAAGCAUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG----------------GGCCUCCAGGGAGUGGAUUUGGAUUCGCUGCGUGGAGGGU .(((((((((((....(((((............)))))..(((....)))..))))))))))).----------------..((((((.(.(((((((....))))))).).)))))).. ( -36.20) >DroYak_CAF1 20780 104 - 1 GAGCGGCGACUAAAAAUAGUUAAGCGUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG----------------GGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGU .(((((((((((....(((((............)))))..(((....)))..))))))))))).----------------..((((((.(.(((((((....))))))).).)))))).. ( -38.40) >DroAna_CAF1 20095 110 - 1 AAA-ACUGACUAAAAAUAGUUAAGCAUCAUAAAAAAAAUACACAAAUUUGCAUAGUUGUUGCCUGGGUCCACUAGGCAGCGGCCUCUAGGGAGUGUAU---------CAGCGUGGAGGGU ...-..((((((....))))))..................(((.....((((..((((((((((((.....))))))))))))(((....))))))).---------....)))...... ( -28.80) >consensus AAGCGGCGACUAAAAAUAGUUAAGCAUCAUAAAAACUACACAAAAAUUUGCAUAGUUGUUGCUG________________GGCCUCCAGGGAGUGGAUUUGGAUCCGCUGCGUGGAGGGU .(((((((((((.((....))..(((......................))).)))))))))))...................((((((.(.(((((((....))))))).).)))))).. (-29.93 = -30.85 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:01 2006