
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,782,096 – 4,782,270 |
| Length | 174 |
| Max. P | 0.838067 |

| Location | 4,782,096 – 4,782,215 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
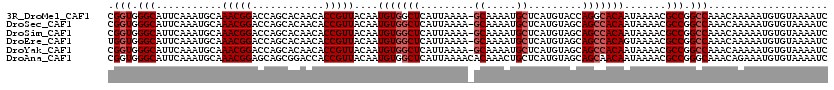
>3R_DroMel_CAF1 4782096 119 - 27905053 CGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA-GCAAAAUGCUCAUGUACCAGGCACAAUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC .(((.(((...........(((((............)))))....((((.((...((..(-((.....)))..))....)).)))).......))).))).................... ( -24.80) >DroSec_CAF1 17401 119 - 1 CGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA-GCAAAAUGCUCAUGUAGCAGCCACAAUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC .(((.(((...........(((((............)))))....((((((((..((..(-((.....)))..))..).))))))).......))).))).................... ( -29.50) >DroSim_CAF1 12823 119 - 1 CGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA-GCAAAAUGCUCAUGUAGCAGCCACAAUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC .(((.(((...........(((((............)))))....((((((((..((..(-((.....)))..))..).))))))).......))).))).................... ( -29.50) >DroEre_CAF1 17605 119 - 1 UGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA-GCAAAAUGCUCAUGUAGCAGCCACAGUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC ((((.(((...........(((((............)))))....((((((((..((..(-((.....)))..))..).))))))).......))).))))................... ( -29.90) >DroYak_CAF1 17793 119 - 1 CGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA-GCAAAAUGCUCAUGUAGCAGCCACAAUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC .(((.(((...........(((((............)))))....((((((((..((..(-((.....)))..))..).))))))).......))).))).................... ( -29.50) >DroAna_CAF1 17255 120 - 1 CGGUGGGCAUUCAAAUGCAAACGGAGCAGCGGACCACCGUUACAAUGUGGCUCAUUAAAACACAAACUGCUCAUGUAGCAGCAACAAUAAAACGCCGGGCAAACAGAAAUGUGUAAAAUC (((((.((((....))))..((((((((((((....)))......((((...........))))..)))))).)))................))))).....(((....)))........ ( -29.00) >consensus CGGUGGGCAUUCAAAUGCAAACGGACCAGCACAACACCGUUACAAUGUGGCUCAUUAAAA_GCAAAAUGCUCAUGUAGCAGCCACAAUAAAACGCCGGCCAAACAAAAAUGUGUAAAAUC .(((.(((...........(((((............)))))....(((((((.........((.....)).........))))))).......))).))).................... (-23.75 = -24.42 + 0.67)


| Location | 4,782,175 – 4,782,270 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -12.37 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4782175 95 + 27905053 ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCGUCC--CACUCACACAC--------------ACACACACAGACU--UGG-----AAGACCCCAACACCAUCAC-ACA-AAU ..(((((((((.(((..((...((((....))))....))..)--))..)))....--------------.............--.((-----......)))))))).....-...-... ( -20.10) >DroSec_CAF1 17480 96 + 1 ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCGUCC--CACUCACACAC--------------ACACACACCCACUU-UGG-----AAGACUCCAACACCAUCAC-ACA-AAU ..(((((((((.(((..((...((((....))))....))..)--))..)))....--------------..............-.((-----(....))))))))).....-...-... ( -21.30) >DroSim_CAF1 12902 94 + 1 ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCGUCC--CACUCACACAC----------------ACACACCCACUU-CGG-----AAGACUCCAACACCAUCAC-ACA-AAU ..(((((((((.(((..((...((((....))))....))..)--))..)))....----------------............-.((-----(....))))))))).....-...-... ( -21.60) >DroEre_CAF1 17684 113 + 1 ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCAUCUCCCACUCACACACACUCACGCUCACACUCACACACCCACUUGUGG-----AAGACCCCAACACCAUCAC-ACG-UUU ..(((((((.(..((((.((..((((....))))..))................................((.((((......)))))-----).)))))))))))).....-...-... ( -21.80) >DroYak_CAF1 17872 105 + 1 ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCGUCCCACACUAACGCACACUC------ACACACACACACCCACUU--GG-----AAGACCCCAACACCAUCAC-ACG-AUU ..(((((((((((((..((...((((....))))....))..))).......))))....------..................--((-----......)))))))).....-...-... ( -21.41) >DroAna_CAF1 17335 101 + 1 ACGGUGGUCCGCUGCUCCGUUUGCAUUUGAAUGCCCACCGCC---AACAUACCGCC----------------ACAGAUACAUUUUCAGAUACAGAUACACGCACACCAUCGCUACGCUAU .(((((((..((((....(((((.(((((((.((.....)).---.......(...----------------...).......))))))).))))).)).))..)))))))......... ( -17.30) >consensus ACGGUGUUGUGCUGGUCCGUUUGCAUUUGAAUGCCCACCGUCC__CACUCACACAC______________ACACACACCCACUU_UGG_____AAGACCCCAACACCAUCAC_ACA_AAU ..(((((((....((((.((..((((....))))..))...........................................(....)........)))).)))))))............. (-12.37 = -12.48 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:51 2006