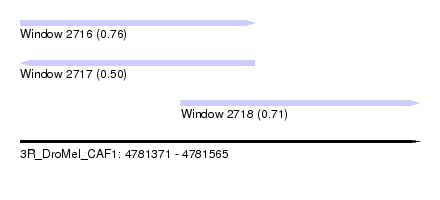
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,781,371 – 4,781,565 |
| Length | 194 |
| Max. P | 0.756349 |

| Location | 4,781,371 – 4,781,485 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4781371 114 + 27905053 CAUACUUCCGCGAUCGGCAAAGCCCGAACG-UUUUUGCCGAAGCCCGGCUUCGCUGUUUCCCCCUCUCUU-CACUAAGCGUCUCU--CUUUUUCUCUUGAAACUCCCUCUCUCGCUCU .........((((((((((((((......)-.))))))))).....((...((((...............-.....)))).....--...((((....))))...))....))))... ( -20.55) >DroSec_CAF1 16763 116 + 1 CAUACUUCCGCGAUCGGCAAAGCCCGAACG-CUUUUGACGAAGCCCGGCUUCGCUGUUUCCCCCACUCUC-CACUGAGCCUCUCUCUCUUCUUCUCUUGGAACUCCCUCUCUCGCUCU .........((((.((((.(((((.(...(-((((....)))))).))))).))))............((-((..(((...............))).))))..........))))... ( -24.26) >DroSim_CAF1 12263 116 + 1 CAUACUUCCGCGAUCGGCAAAGCCCGAACG-CUUUUGCCGAAGCCCGGCUUCGCUGUUUCCCCCACUCUC-CACUAAGCCUCUCUUUCUACUUCUCUUGGAACUCCCUCUCUCGCUCU .....((((((..((((((((((......)-).)))))))).))..(((((.(.((..............-))).)))))..................))))................ ( -23.84) >DroEre_CAF1 17023 105 + 1 CAUACUUCCGCGAUCGGCAAAACCCGAACGGCAUUUGCCGAAGCCCGGCUUCGCUGUUUCCCCU-CUCUCACU------------CUCUCCCUCUCCUGCAACUCCCUCUCUCGCUCU .........((((((((......))))(((((....((((.....))))...))))).......-........------------..........................))))... ( -21.30) >consensus CAUACUUCCGCGAUCGGCAAAGCCCGAACG_CUUUUGCCGAAGCCCGGCUUCGCUGUUUCCCCCACUCUC_CACUAAGCCUCUCUCUCUUCUUCUCUUGGAACUCCCUCUCUCGCUCU .........((((((((......)))).........(.((((((...))))))).........................................................))))... (-15.60 = -15.60 + -0.00)

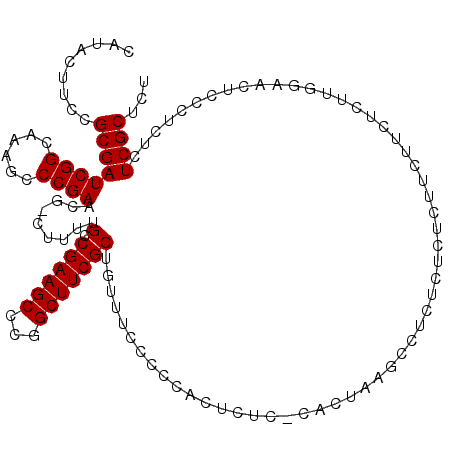
| Location | 4,781,371 – 4,781,485 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.05 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4781371 114 - 27905053 AGAGCGAGAGAGGGAGUUUCAAGAGAAAAAG--AGAGACGCUUAGUG-AAGAGAGGGGGAAACAGCGAAGCCGGGCUUCGGCAAAAA-CGUUCGGGCUUUGCCGAUCGCGGAAGUAUG ...((((..(.(((.(((((...........--.))))).)))....-.........(....).((((((((..((....)).....-(....))))))))))..))))......... ( -29.10) >DroSec_CAF1 16763 116 - 1 AGAGCGAGAGAGGGAGUUCCAAGAGAAGAAGAGAGAGAGGCUCAGUG-GAGAGUGGGGGAAACAGCGAAGCCGGGCUUCGUCAAAAG-CGUUCGGGCUUUGCCGAUCGCGGAAGUAUG ..............(.((((............((..(((((((.((.-....((...(....).))...)).))))))).))....(-((.((((......)))).))))))).)... ( -32.00) >DroSim_CAF1 12263 116 - 1 AGAGCGAGAGAGGGAGUUCCAAGAGAAGUAGAAAGAGAGGCUUAGUG-GAGAGUGGGGGAAACAGCGAAGCCGGGCUUCGGCAAAAG-CGUUCGGGCUUUGCCGAUCGCGGAAGUAUG ...((((.....((....))................((((((..((.-....((...(....).))...))..))))))((((((.(-(......))))))))..))))......... ( -32.90) >DroEre_CAF1 17023 105 - 1 AGAGCGAGAGAGGGAGUUGCAGGAGAGGGAGAG------------AGUGAGAG-AGGGGAAACAGCGAAGCCGGGCUUCGGCAAAUGCCGUUCGGGUUUUGCCGAUCGCGGAAGUAUG ...((((.....((..((((.............------------..(....)-...(....).))))..)).....((((((((.(((.....)))))))))))))))......... ( -29.00) >consensus AGAGCGAGAGAGGGAGUUCCAAGAGAAGAAGAGAGAGAGGCUUAGUG_GAGAGUGGGGGAAACAGCGAAGCCGGGCUUCGGCAAAAG_CGUUCGGGCUUUGCCGAUCGCGGAAGUAUG ...((((..(...............................................(....).((((((((.((...((........)).)).)))))))))..))))......... (-23.24 = -23.05 + -0.19)



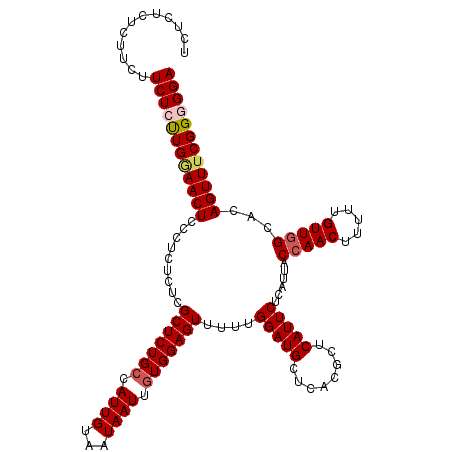
| Location | 4,781,449 – 4,781,565 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.34 |
| Energy contribution | -23.28 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4781449 116 + 27905053 UCUCU--CUUUUUCUCUUGAAACUCCCUCUCUCGCUCUGGCAUUGUAAUAAUUGUGGAGUUUUUGGAUGCUCACGCUCAUUCUCAUUACCAACUUUUUGUUGGCACAGUUUCGGGGGA .....--.....(((((((((((..........((....))...........(((((((......((.((....))))...)))....(((((.....)))))))))))))))))))) ( -28.10) >DroSec_CAF1 16841 118 + 1 UCUCUCUCUUCUUCUCUUGGAACUCCCUCUCUCGCUCUGCCAUUGUAAUAAUUGUGGAGUUUUUGGAUGCUCACGCUCAUUCUCAUUACCAACUUUUUGUUGGCACAGUUUCGGGGGA ............((((((((((((.........((((..(.((((...)))).)..))))....(((((........)))))......(((((.....)))))...)))))))))))) ( -27.50) >DroSim_CAF1 12341 117 + 1 UCUCUUUCUACUUCUCUUGGAACUCCCUCUCUCGCUCUGCCAUUGUAAUAAUUGUGGAGUUUUUGGAUGCUCACGCUCAUUCUCAUUACAAACUUUUUGUUGGCACAGUUUCGG-GGA .....(((((.......))))).(((((.....(((.(((((((((((((((.(((((((........)))).)))..)))...)))))))((.....))))))).)))...))-))) ( -24.90) >DroEre_CAF1 17095 113 + 1 -----CUCUCCCUCUCCUGCAACUCCCUCUCUCGCUCUGCCAUUGUAAUAAUUGUGGAGUUUUUGGAUGCUCACGCUCAUUCUCAUUACCAACUUUUUGUUGGCACAGUUUCGGUGGA -----..................................((((((....((((((((((......((.((....))))...)))....(((((.....)))))))))))).)))))). ( -23.40) >consensus UCUCUCUCUUCUUCUCUUGGAACUCCCUCUCUCGCUCUGCCAUUGUAAUAAUUGUGGAGUUUUUGGAUGCUCACGCUCAUUCUCAUUACCAACUUUUUGUUGGCACAGUUUCGGGGGA ............((((((((((((.........(((((((.((((...)))).)))))))....(((((........)))))......(((((.....)))))...)))))))))))) (-22.34 = -23.28 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:49 2006