
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,777,801 – 4,777,961 |
| Length | 160 |
| Max. P | 0.993967 |

| Location | 4,777,801 – 4,777,921 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -51.78 |
| Consensus MFE | -48.60 |
| Energy contribution | -48.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4777801 120 + 27905053 GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGAAAGGCCUUGGCGCAGACAUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGA .............(((((..(((.((((((((.((((((.((((....))))....))))..)).))))))))(((((.(..((((.....))))...).))))))))..)))))..... ( -51.50) >DroSec_CAF1 13290 120 + 1 GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGA .............(((((..(((.((((((((.((((((.((((....))))....))))..)).))))))))(((((.(..((((.....))))...).))))))))..)))))..... ( -51.10) >DroEre_CAF1 13448 120 + 1 GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAGGUGCACCUUGA .............((((((((((.((((((((.((((((.((((....))))....))))..)).))))))))(((((.(..((((.....))))...).)))))))))))))))..... ( -56.30) >DroYak_CAF1 13602 120 + 1 GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGA .............(((((..(((.((((((((.((((((.((((....))))....))))..)).))))))))(((((.(..((((.....))))...).))))))))..)))))..... ( -51.10) >DroAna_CAF1 13036 120 + 1 GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGGAAGGCCUUGGCGCAGACGUCGCAGCGGAAGGGCUUCUCGCCCGUGUGGGUGCGAAGGUGCACCUUGA ..........((((..((((....))))..)))).((((((((((((.((((..(((((((..(((..........)))..)))))))..)))).((......))))))))))))))... ( -48.90) >consensus GCCCAAUUCUCUUGUGUAUCUGUUGAUGUUUGAGCAGGUGCGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGA .............((((((.(((.((((((((.((((((.((((....))))....))))..)).))))))))(((((.(..((((.....))))...).)))))))).))))))..... (-48.60 = -48.84 + 0.24)


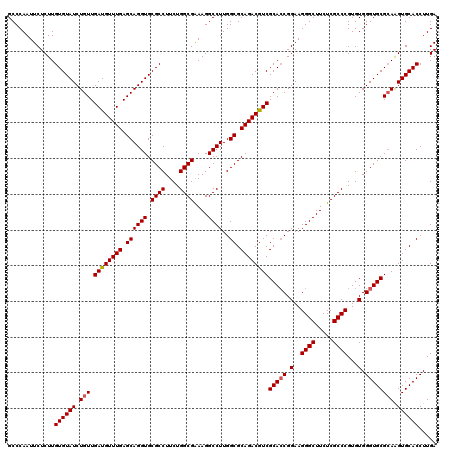
| Location | 4,777,801 – 4,777,921 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -41.02 |
| Energy contribution | -41.50 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4777801 120 - 27905053 UCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGAUGUCUGCGCCAAGGCCUUUCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.((((((((((.....((((.....))))....))))))..(....)..))))((((((.....))))))))))))((((((...((.............))...)))))) ( -42.82) >DroSec_CAF1 13290 120 - 1 UCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.((((((((((.....((((.....))))....)))))).((....)).))))((((((.....))))))))))))((((((...((.............))...)))))) ( -44.42) >DroEre_CAF1 13448 120 - 1 UCAAGGUGCACCUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.(((.((........))(((((((.(((.....((((((.....))))))..))).)))))))...))).))))))((((((...((.............))...)))))) ( -43.42) >DroYak_CAF1 13602 120 - 1 UCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.((((((((((.....((((.....))))....)))))).((....)).))))((((((.....))))))))))))((((((...((.............))...)))))) ( -44.42) >DroAna_CAF1 13036 120 - 1 UCAAGGUGCACCUUCGCACCCACACGGGCGAGAAGCCCUUCCGCUGCGACGUCUGCGCCAAGGCCUUCCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.((((((......)...((((.((((.((((..((.(((.....))))).)))).)))))))).))))).))))))((((((...((.............))...)))))) ( -42.52) >consensus UCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCGCACCUGCUCAAACAUCAACAGAUACACAAGAGAAUUGGGC ...((((((.((((((((((.....((((.....))))....)))))).((....)).))))((((((.....))))))))))))((((((...((.............))...)))))) (-41.02 = -41.50 + 0.48)

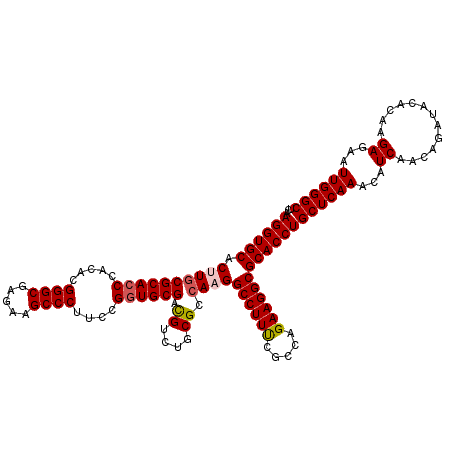
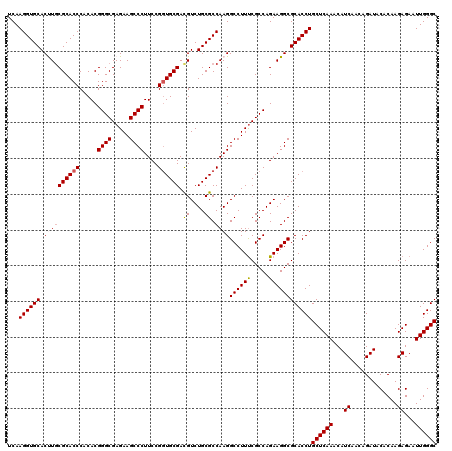
| Location | 4,777,841 – 4,777,961 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -52.60 |
| Consensus MFE | -47.24 |
| Energy contribution | -47.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4777841 120 + 27905053 CGCCUUCUGGCGAAAGGCCUUGGCGCAGACAUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGAUGUUGGACUUGUUGAGGAAUCCGCGAUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((((.(..((((.....))))...).)))))((((.(((.(((..(((.......)))..))).....))).)))).)))))))). ( -53.00) >DroSec_CAF1 13330 120 + 1 CGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGAUGUUGGACUUGUUGAGGAAUCCGCGAUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((((.(..((((.....))))...).)))))((((.(((.(((..(((.......)))..))).....))).)))).)))))))). ( -52.60) >DroEre_CAF1 13488 120 + 1 CGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAGGUGCACCUUGAUGUUGGACUUGUUGAGGAAUCCCCGAUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((.(((..((..(((((((((....))))..((((((.((.(.....).)).)))))).)))))..)))))..))).)))))))). ( -49.60) >DroYak_CAF1 13642 120 + 1 CGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGAUGUUGGACUUGUUGAGGAAGCCCCGAUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((.(((...((((((((((((....))))).((((((.((.(.....).)).))))))...))))))))))..))).)))))))). ( -51.90) >DroAna_CAF1 13076 120 + 1 CGCCUUCUGGCGGAAGGCCUUGGCGCAGACGUCGCAGCGGAAGGGCUUCUCGCCCGUGUGGGUGCGAAGGUGCACCUUGAUAUUGGACUUGUUGAGGAACCCCCUGUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((((((..(((((((.(((((....))))).)))).....(((..(((.......)))..)))..))).))))))).)))))))). ( -55.90) >consensus CGCCUUCUGGCGAAAGGCCUUGGCGCAGACGUCGCACCGGAAGGGCUUCUCGCCCGUGUGGGUGCGCAAGUGCACCUUGAUGUUGGACUUGUUGAGGAAUCCCCGAUUGCAGAUGCUGCA .(((....(((.....)))..)))((((.(((((((((.(..((((.....))))...).))))).....(((.(((..(((.......)))..)))((((...))))))))))))))). (-47.24 = -47.28 + 0.04)



| Location | 4,777,841 – 4,777,961 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -42.82 |
| Energy contribution | -43.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4777841 120 - 27905053 UGCAGCAUCUGCAAUCGCGGAUUCCUCAACAAGUCCAACAUCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGAUGUCUGCGCCAAGGCCUUUCGCCAGAAGGCG .((((((((.((((..(((((((........))))).((......))))..))))(((((.....((((.....))))....))))))))).))))(((..(((.....)))....))). ( -45.30) >DroSec_CAF1 13330 120 - 1 UGCAGCAUCUGCAAUCGCGGAUUCCUCAACAAGUCCAACAUCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCG (((((...))))).....(((((........)))))........((((((..((((((((.....((((.....))))....)))))).))..))))))...((((((.....)))))). ( -47.20) >DroEre_CAF1 13488 120 - 1 UGCAGCAUCUGCAAUCGGGGAUUCCUCAACAAGUCCAACAUCAAGGUGCACCUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCG (((((...))))).....(((((........)))))........((((((..((((((((.....((((.....))))....)))))).))..))))))...((((((.....)))))). ( -46.10) >DroYak_CAF1 13642 120 - 1 UGCAGCAUCUGCAAUCGGGGCUUCCUCAACAAGUCCAACAUCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCG (((((...)))))....((((((.......))))))........((((((..((((((((.....((((.....))))....)))))).))..))))))...((((((.....)))))). ( -49.10) >DroAna_CAF1 13076 120 - 1 UGCAGCAUCUGCAACAGGGGGUUCCUCAACAAGUCCAAUAUCAAGGUGCACCUUCGCACCCACACGGGCGAGAAGCCCUUCCGCUGCGACGUCUGCGCCAAGGCCUUCCGCCAGAAGGCG .(((((.((.(((.(.(((((((.(((.....((((........(((((......))))).....))))))).)))))))..).))))).).))))......((((((.....)))))). ( -41.72) >consensus UGCAGCAUCUGCAAUCGGGGAUUCCUCAACAAGUCCAACAUCAAGGUGCACUUGCGCACCCACACGGGCGAGAAGCCCUUCCGGUGCGACGUCUGCGCCAAGGCCUUUCGCCAGAAGGCG (((((...))))).....(((((........)))))........((((((..((((((((.....((((.....))))....)))))).))..))))))...((((((.....)))))). (-42.82 = -43.02 + 0.20)


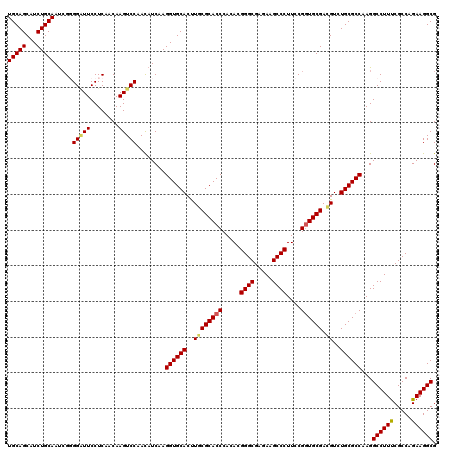
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:42 2006