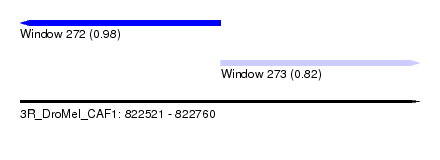
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 822,521 – 822,760 |
| Length | 239 |
| Max. P | 0.976561 |

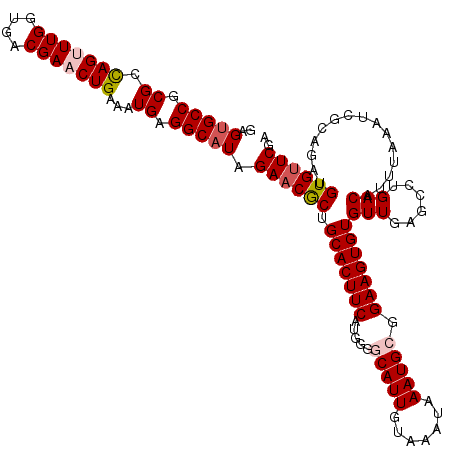
| Location | 822,521 – 822,641 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -34.16 |
| Energy contribution | -35.04 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 822521 120 - 27905053 GAGUGCCUCGCCAGUUUGGUGACGAACUGAAAUGAGGCAUAGAACGCUGCACUUCAUGGGGCAUUGUAAAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGUUCGA ..((((((((.(((((((....)))))))...)))))))).((((((.(((((((.....(((((.......))))).)))))))(((......)))...............)))))).. ( -44.10) >DroSec_CAF1 4994 120 - 1 GAGUGCCGCGCCAGUUUGGUGACGAACUGAAAUGAGGCAUAGAACGCUGCACUUCAUGGGGCAUUGUAAAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGUUCGA ..(((((.((.(((((((....)))))))...)).))))).((((((.(((((((.....(((((.......))))).)))))))(((......)))...............)))))).. ( -39.70) >DroSim_CAF1 2173 120 - 1 GAGUGCCGCGCCAGUUUGGUGACGAACUGAAAUGAGGCAUAGAACGCUGCACUUCAUGGGGCAUUGUAAAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGUUCGA ..(((((.((.(((((((....)))))))...)).))))).((((((.(((((((.....(((((.......))))).)))))))(((......)))...............)))))).. ( -39.70) >DroEre_CAF1 5764 119 - 1 GAGUGCCGCGCCAGCUUGGUGACGAUCUGAAAUGAGGCAUAGAACGCUGCACUUCAUGGG-CAUUGUAGAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGGUCGA ....((((((.(((.(((....))).)))(((((((((..((....))(((((((....(-((((.......))))).))))))).....))))..)))))..........))))))... ( -33.60) >DroYak_CAF1 5420 120 - 1 AAGAGCCGCGCUAGUUUGGUGACGAUCUGAAAUGAGGCAUAGAACACUGCACUUCAUGGGACAUUGUAAAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGUUCGA ....(((.((.(((.(((....))).)))...)).)))...((((((.(((((((....(.((((.......))))).)))))))(((......)))...............)))))).. ( -29.00) >consensus GAGUGCCGCGCCAGUUUGGUGACGAACUGAAAUGAGGCAUAGAACGCUGCACUUCAUGGGGCAUUGUAAAUAAAUGCGGAAGUGUGUUGAGCCUGACAUUUAAAUCGCAGAUGUGUUCGA ..(((((.((.(((((((....)))))))...)).))))).((((((.(((((((.....(((((.......))))).)))))))(((......)))...............)))))).. (-34.16 = -35.04 + 0.88)

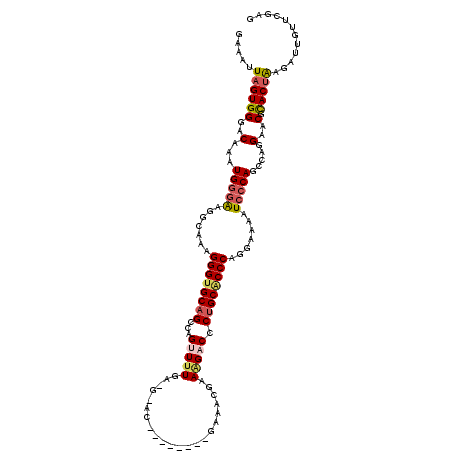
| Location | 822,641 – 822,760 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 822641 119 + 27905053 GAAAUUAGUGGGACAAAUGUGAAGGAAAAGGGCGCAGCCAGUUCUGA-GCACCGAGGCUCGAAGCGAAAGACCCUGCACCCAGAAAAAUCCCAGCCAGGAACGCACCAAGCUUGGUCGAG ........(((((((....))........(((.((((....(((.((-((......))))))).(....)...)))).))).......)))))((((((...........)))))).... ( -30.70) >DroSec_CAF1 5114 105 + 1 GAAAUUAGUGGGACAAAUGGGAAGGCAAAGGGUGCAGCCAGCUUU---------------GAAACGAAAGACCCUGCGCCCAGGUAAAUCCCAGCCAGGAACGCACUAAGAGUGUUCGAG ..........((.....(((((..((...((((((((....((((---------------......))))...))))))))..))...))))).))..((((((.......))))))... ( -34.70) >DroSim_CAF1 2293 104 + 1 GAAAUUAGUGGGACAAAUGGGAAGGCAAAGGGUGCAGCCAGCUUU---------------GAAACGAAAGACCCUGCACCCACGAAAAUCCCAGCCAGGAACGCACUAA-AGUGUUCGAG ..........((.....(((((.......((((((((....((((---------------......))))...)))))))).......))))).))..((((((.....-.))))))... ( -33.34) >DroEre_CAF1 5883 108 + 1 AAAGUUAGUGGGACAAAUGGGGAGGCAAGGGGUGCAGCCAGUUUUGAAGGAC------------CGCAGGACCCCGCACCCAGGAAAGUCCCAACUAGGAACGUACUGCGUUUACUUGAG .(((((((((((((...((((...((..(((((.(.((..((((....))))------------.)).).))))))).)))).....))))).)))).(((((.....)))))))))... ( -38.60) >DroYak_CAF1 5540 118 + 1 GAAAUUAGUGGAACAAAUGGAAGAGCAACGGGUGCAGCCAGUUUUGA-GCACCGAGGCUCGACACGAAAGACCCUGCACCCAG-AAAAUCCCAACCAGGAACGUACUGCGUUUAUUUGAG .............((((((((...(((((((((((((......((((-((......))))))..(....)...))))))))..-....(((......)))..))..))).)))))))).. ( -36.00) >consensus GAAAUUAGUGGGACAAAUGGGAAGGCAAAGGGUGCAGCCAGUUUUGA_G_AC________GAAACGAAAGACCCUGCACCCAGGAAAAUCCCAGCCAGGAACGCACUAAGAUUGUUCGAG .....((((((..(...(((((.......((((((((...(((((......................))))).)))))))).......))))).....)..).)))))............ (-19.25 = -19.97 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:28 2006