
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,771,551 – 4,771,682 |
| Length | 131 |
| Max. P | 0.979910 |

| Location | 4,771,551 – 4,771,660 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -24.31 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
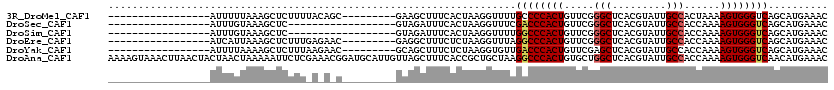
>3R_DroMel_CAF1 4771551 109 + 27905053 CUGUGGCUCCUGCCGCUUUUU---UACUUUUUUCUGGCUUGUUUCAUGCUGACCCACUUUUAGUGGCAAUACGUGAGCCCGAACAGUGGGGCAAAACCUUAGUGAAAGCUUC-------- ..(((((....))))).((((---((((.......((......(((((.....((((.....)))).....)))))((((........))))....))..))))))))....-------- ( -29.70) >DroSec_CAF1 6943 109 + 1 CUGUGGCUGCUGCCGCUUUUU---UACUUUUUUCUGGCUUGUUUCAUGCUGACCCACUUUUGGUGGCAAUACGUGAGCCCGAACAGUGGGUCGAAACCUUAGUGAAAUCUAC-------- ..(((((....)))))..(((---((((..((((.(((.........)))((((((((.((.(.(((.........)))).)).))))))))))))....))))))).....-------- ( -36.60) >DroSim_CAF1 5382 109 + 1 CUGUGGCUGCUGCCGCUUUUU---UACUUUUUUCUGGCUUGUUUCAUGCUGACCCACUUUUGGUGGCAAUACGUGAGCCCGAACAGUGGGCCAAAACCUUAGUGAAAUCUAC-------- ..(((((....)))))..(((---((((......(((((..((((..(((.((((((.....))))......)).)))..)))..)..))))).......))))))).....-------- ( -31.62) >DroAna_CAF1 6807 120 + 1 CUGCGGCUGCUGCCGCUUUUUUCCUACUCUUUUCUGGCUUGUUUCAUGUUGACCCACUUUUGGUGGCAAUACGUGAGCCAGCACAGUGGGCCUUAGCAGCGGUGAAAGCUAACAAUGCAU .(((((((..(((((((.((..((((((.....(((((.....((((((....((((.....))))....)))))))))))...))))))....)).)))))))..))))......))). ( -40.20) >consensus CUGUGGCUGCUGCCGCUUUUU___UACUUUUUUCUGGCUUGUUUCAUGCUGACCCACUUUUGGUGGCAAUACGUGAGCCCGAACAGUGGGCCAAAACCUUAGUGAAAGCUAC________ ..(((((....)))))........((((......(((((..((((..(((.((((((.....))))......)).)))..)))..)..))))).......))))................ (-24.31 = -24.25 + -0.06)

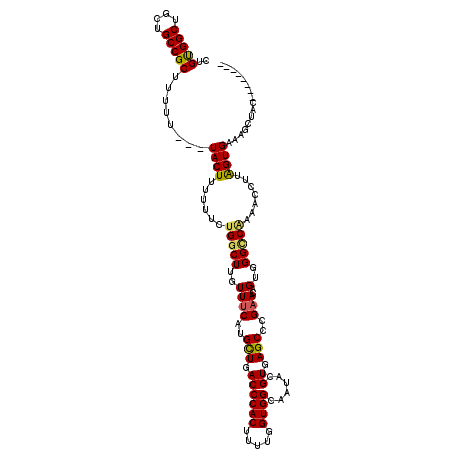

| Location | 4,771,588 – 4,771,682 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4771588 94 - 27905053 -----------------AUUUUUAAAGCUCUUUUACAGC---------GAAGCUUUCACUAAGGUUUUGCCCCACUGUUCGGGCUCACGUAUUGCCACUAAAAGUGGGUCAGCAUGAAAC -----------------.........(((........((---------((((((((....))))))))))((((((.....(((.........)))......))))))..)))....... ( -26.40) >DroSec_CAF1 6980 85 - 1 -----------------AUUUGUAAAGCUC------------------GUAGAUUUCACUAAGGUUUCGACCCACUGUUCGGGCUCACGUAUUGCCACCAAAAGUGGGUCAGCAUGAAAC -----------------.........(((.------------------...((..((.....))..))((((((((.....(((.........)))......)))))))))))....... ( -24.10) >DroSim_CAF1 5419 85 - 1 -----------------AUUUGUAAAGCUC------------------GUAGAUUUCACUAAGGUUUUGGCCCACUGUUCGGGCUCACGUAUUGCCACCAAAAGUGGGUCAGCAUGAAAC -----------------...(((((((((.------------------.(((......))).))))))((((((((.....(((.........)))......)))))))).)))...... ( -23.60) >DroEre_CAF1 7194 94 - 1 -----------------AUCAUUAAAGCUCUUUGAGAAC---------GAGGCUUUCUCUAAGGUUUAGGCCCACUGUUCGGGCUCACGUAUUGCCACCAAAAGUGGGUCAGCAUGAAAC -----------------.((((..((((.(((.(((((.---------......))))).))))))).((((((((.....(((.........)))......))))))))...))))... ( -29.10) >DroYak_CAF1 7131 94 - 1 -----------------AUUUUAAAAGCUCUUUAAGAAC---------GCAGCUUUCUCUAAGGUGUUGACCCACUGUUCGAGCUCACGUAUUGCCACCAAAAGUGGGUCAGCAUGAAAC -----------------......(((((((.........---------).)))))).......(((((((((((((......((.........)).......)))))))))))))..... ( -25.92) >DroAna_CAF1 6847 120 - 1 AAAAGUAAACUUAACUACUAACUAAAAAUUCUCGAAACGGAUGCAUUGUUAGCUUUCACCGCUGCUAAGGCCCACUGUGCUGGCUCACGUAUUGCCACCAAAAGUGGGUCAACAUGAAAC ...((((..........(((((.....((((.......)))).....)))))..........))))..((((((((.((.((((.........)))).))..)))))))).......... ( -27.75) >consensus _________________AUUUUUAAAGCUCUUU_A_A_C_________GUAGCUUUCACUAAGGUUUUGGCCCACUGUUCGGGCUCACGUAUUGCCACCAAAAGUGGGUCAGCAUGAAAC ....................................................................((((((((.....(((.........)))......)))))))).......... (-17.82 = -17.98 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:27 2006