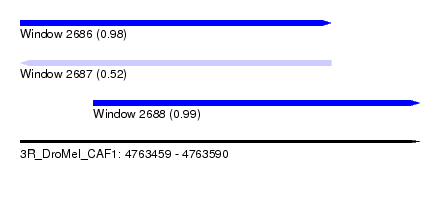
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,763,459 – 4,763,590 |
| Length | 131 |
| Max. P | 0.987220 |

| Location | 4,763,459 – 4,763,561 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.77 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
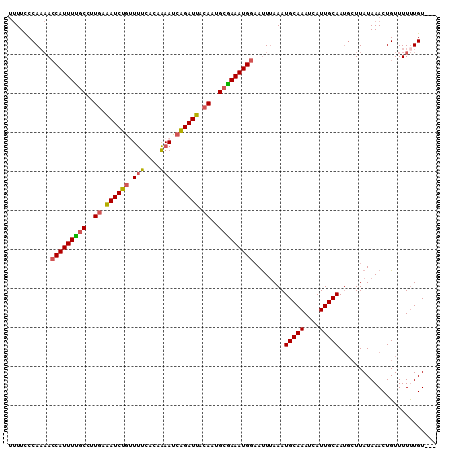
>3R_DroMel_CAF1 4763459 102 + 27905053 AAACUUCUCUUCAAACUUUUCCGCUUUUCCCAAAACCAUUUUGCUUUGAAAUCUGUUUUUACCAAAUCAGAUUAAACUGCAAAAUGGAACUUAAAUGCAAAU ......................((...........(((((((((.....((((((.(((....))).)))))).....))))))))).........)).... ( -17.95) >DroSec_CAF1 2652 102 + 1 AAACUUCUCUUCAAACUUUUCCGCUUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUUACAAAAUCAGAUUACACUGCGAAAUGGAAUUUAAAUGCAAAU ......................((...........(((((((((..((.((((((.(((....))).)))))).))..))))))))).........)).... ( -19.55) >DroSim_CAF1 2654 102 + 1 AAACUUCUCUUCAAACUUUUCCGCUUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUUACAAAAUCAGAUUACACCGCGAAAUGGAAUUUAAAUGCAAAU ......................((...........(((((((((..((.((((((.(((....))).)))))).))..))))))))).........)).... ( -19.45) >DroEre_CAF1 2603 101 + 1 AAACUUUUCCUCAAACUUUUCCGCUUUUCCCA-AACCAUUUUGCCUUGAGAUCUGUUUUCACAAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAU ......................((........-..(((((((((.(((.((((((.(((....))).)))))).))).))))))))).........)).... ( -22.01) >DroYak_CAF1 2646 102 + 1 AAACUUUUCCUAAAACUUUUCCGCUUUUCCCAAAACCAUUUUGCCUUGAGAUCUGCUUUCACCAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAU ......................((...........(((((((((.(((.((((((.(((....))).)))))).))).))))))))).........)).... ( -21.25) >DroPer_CAF1 2623 100 + 1 AAACUUGUCUAAAAAUGCUCUUGAAAUUUCCAACA-CAUUUCCCAUUGCAAUCUAAUCCCAUUGAAU-GGAUUGCAAAGCGAAAUGAUAUUCAAAUGCAAAU ...............(((..(((((..........-((((((.(.((((((((((.((.....)).)-))))))))).).))))))...)))))..)))... ( -22.62) >consensus AAACUUCUCUUCAAACUUUUCCGCUUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUCACAAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAU ...................................(((((((((..((.((((((.(((....))).)))))).))..)))))))))............... (-16.85 = -16.77 + -0.08)


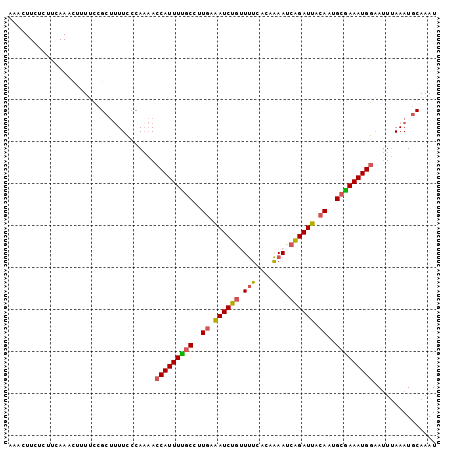
| Location | 4,763,459 – 4,763,561 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4763459 102 - 27905053 AUUUGCAUUUAAGUUCCAUUUUGCAGUUUAAUCUGAUUUGGUAAAAACAGAUUUCAAAGCAAAAUGGUUUUGGGAAAAGCGGAAAAGUUUGAAGAGAAGUUU .(((((.(((.((..(((((((((.....((((((.(((....))).)))))).....)))))))))..))...))).)))))................... ( -23.90) >DroSec_CAF1 2652 102 - 1 AUUUGCAUUUAAAUUCCAUUUCGCAGUGUAAUCUGAUUUUGUAAAAACAGAUUUCAAGGCAAAAUGGUUUUGGGAAAAGCGGAAAAGUUUGAAGAGAAGUUU .(((((.(((.((..((((((.((..((.((((((.(((....))).)))))).))..)).))))))..))...))).)))))................... ( -22.70) >DroSim_CAF1 2654 102 - 1 AUUUGCAUUUAAAUUCCAUUUCGCGGUGUAAUCUGAUUUUGUAAAAACAGAUUUCAAGGCAAAAUGGUUUUGGGAAAAGCGGAAAAGUUUGAAGAGAAGUUU .(((((.(((.((..((((((.((..((.((((((.(((....))).)))))).))..)).))))))..))...))).)))))................... ( -22.00) >DroEre_CAF1 2603 101 - 1 AUUUGCAUUUAAAUUCCAUUUCGCAUUGUAAUCUGAUUUUGUGAAAACAGAUCUCAAGGCAAAAUGGUU-UGGGAAAAGCGGAAAAGUUUGAGGAAAAGUUU ((((...((((((((...((((((.............(((((....)))))(((((((.(.....).))-)))))...)))))).))))))))...)))).. ( -21.90) >DroYak_CAF1 2646 102 - 1 AUUUGCAUUUAAAUUCCAUUUCGCAUUGUAAUCUGAUUUGGUGAAAGCAGAUCUCAAGGCAAAAUGGUUUUGGGAAAAGCGGAAAAGUUUUAGGAAAAGUUU .(((((.(((.((..((((((.((.(((..(((((.(((.....))))))))..))).)).))))))..))...))).)))))................... ( -22.40) >DroPer_CAF1 2623 100 - 1 AUUUGCAUUUGAAUAUCAUUUCGCUUUGCAAUCC-AUUCAAUGGGAUUAGAUUGCAAUGGGAAAUG-UGUUGGAAAUUUCAAGAGCAUUUUUAGACAAGUUU ...(((.((((((...((((((.(.((((((((.-((((....))))..)))))))).).))))))-..........)))))).)))............... ( -23.82) >consensus AUUUGCAUUUAAAUUCCAUUUCGCAGUGUAAUCUGAUUUUGUAAAAACAGAUUUCAAGGCAAAAUGGUUUUGGGAAAAGCGGAAAAGUUUGAAGAGAAGUUU .(((((.(((.((..((((((.((..((.((((((.(((....))).)))))).))..)).))))))..))...))).)))))................... (-16.19 = -16.92 + 0.73)



| Location | 4,763,483 – 4,763,590 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.57 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
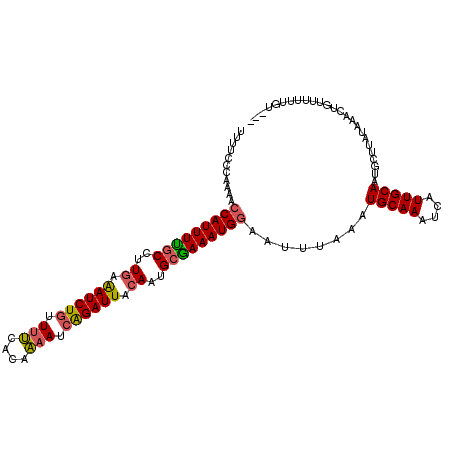
>3R_DroMel_CAF1 4763483 107 + 27905053 UUUUCCCAAAACCAUUUUGCUUUGAAAUCUGUUUUUACCAAAUCAGAUUAAACUGCAAAAUGGAACUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU--- ......((((((((((((((.....((((((.(((....))).)))))).....)))))))))........(((((....)))))................))))).--- ( -22.80) >DroSec_CAF1 2676 107 + 1 UUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUUACAAAAUCAGAUUACACUGCGAAAUGGAAUUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU--- ......((((((((((((((..((.((((((.(((....))).)))))).))..)))))))))........(((((....)))))................))))).--- ( -24.40) >DroSim_CAF1 2678 107 + 1 UUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUUACAAAAUCAGAUUACACCGCGAAAUGGAAUUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU--- ......((((((((((((((..((.((((((.(((....))).)))))).))..)))))))))........(((((....)))))................))))).--- ( -24.30) >DroEre_CAF1 2627 106 + 1 UUUUCCCA-AACCAUUUUGCCUUGAGAUCUGUUUUCACAAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU--- ......((-(((((((((((.(((.((((((.(((....))).)))))).))).)))))))))........(((((....))))).................)))).--- ( -25.40) >DroYak_CAF1 2670 107 + 1 UUUUCCCAAAACCAUUUUGCCUUGAGAUCUGCUUUCACCAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU--- ......((((((((((((((.(((.((((((.(((....))).)))))).))).)))))))))........(((((....)))))................))))).--- ( -26.10) >DroPer_CAF1 2647 108 + 1 AAUUUCCAACA-CAUUUCCCAUUGCAAUCUAAUCCCAUUGAAU-GGAUUGCAAAGCGAAAUGAUAUUCAAAUGCAAAUCAUUGCAAAUGUUAUAAACUGUUUUCUGUUAC .......((((-((((((.(.((((((((((.((.....)).)-))))))))).).)))))).........(((((....)))))..))))................... ( -24.50) >consensus UUUUCCCAAAACCAUUUUGCCUUGAAAUCUGUUUUCACAAAAUCAGAUUACAAUGCGAAAUGGAAUUUAAAUGCAAAUCAUUGCAAUGCUUAUAAACUGUUUUUUGU___ ...........(((((((((..((.((((((.(((....))).)))))).))..)))))))))........(((((....)))))......................... (-20.65 = -20.57 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:20 2006