
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,723,502 – 4,723,598 |
| Length | 96 |
| Max. P | 0.891589 |

| Location | 4,723,502 – 4,723,598 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -11.63 |
| Energy contribution | -12.83 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4723502 96 + 27905053 AAUG-------------------UGGGAGA-AGUGAUUGUGGUCGAAAUCAUAUGGCCCAUUCAUUUGGAGUAAUGAAAACAAUUGAAAGAAAGCCAGCCAGCAUCUUAAUGGCAA ..((-------------------((((..(-.((((((........)))))).)..))).((((((......)))))).)))...............((((.........)))).. ( -19.80) >DroSec_CAF1 67372 98 + 1 AAUGUUUUCGCUUCG------AGUGGGAGA-AGUGAUUGUUGCCGAAAUCAUAUGUCCCAUUCAAUUGGAGUAAUGAAAACAAUUGGAAGAAAGCCAGCCAG-----------CAA ..(((((((((((((------(((((((.(-.((((((........)))))).).))))))))....)))))...))))))).((((..(.....)..))))-----------... ( -29.80) >DroSim_CAF1 70879 98 + 1 AAUGUUUUCGCUUCG------AGUGGGAGA-AGUGAUUGUGGUCGAAAUCAUAUGGCCCAUUCAAUUGGAGUAAUGAAAACAAUUGGAAGAAAGCCAGCCAG-----------CAA ..(((((((((((((------((((((..(-.((((((........)))))).)..)))))))....)))))...))))))).((((..(.....)..))))-----------... ( -26.90) >DroEre_CAF1 71255 102 + 1 AAUGUUUUUCUUUUG------AGUGGGAGA-AGUGAUUGUGGUCGAAAUCAUAUGGCCCAUUCAACUGGAGUAAUGAAAACAAUU---AGAAAGCCAGC----AUCCUAAUGACAA ..((((......(((------((((((..(-.((((((........)))))).)..)))))))))((((..(..(((......))---)..)..)))).----........)))). ( -24.20) >DroYak_CAF1 70833 102 + 1 AAUGUUUUCCUUUUG------AGUGGGAGA-AGUGAUUGUGGUCGAAAUCAUAUGGCCCAUUCAAUUGGAGUAAUGAAAACAAUU---AGAAAGCCAGC----AUCUUAAUGGCAA ..((((((((..(((------((((((..(-.((((((........)))))).)..)))))))))..))).......)))))...---.....((((..----.......)))).. ( -30.11) >DroPer_CAF1 94510 114 + 1 AAUGUUUUUCUUACGAGGUACAGGGGAAGAAACUGAUUGUAAGGGAGAUCUUAUUACCCUUCCAAUUGGUCUAAUGAGAGCCAUUCA--CAGAGGCAGGAGAAAUGCAUGUGGGUA ....(((..(((((((....(((.........))).)))))))..)))((((((((.((........))..))))))))...(((((--((...(((.......))).))))))). ( -29.70) >consensus AAUGUUUUCCUUUCG______AGUGGGAGA_AGUGAUUGUGGUCGAAAUCAUAUGGCCCAUUCAAUUGGAGUAAUGAAAACAAUUG__AGAAAGCCAGCCAG_AUC_UAAUGGCAA ..(((((((............((((((.....((((((........))))))....)))))).............))))))).................................. (-11.63 = -12.83 + 1.20)

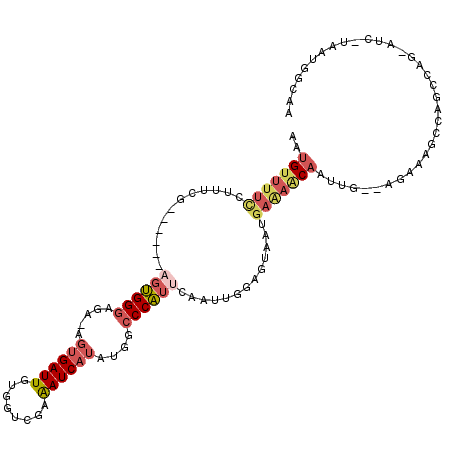
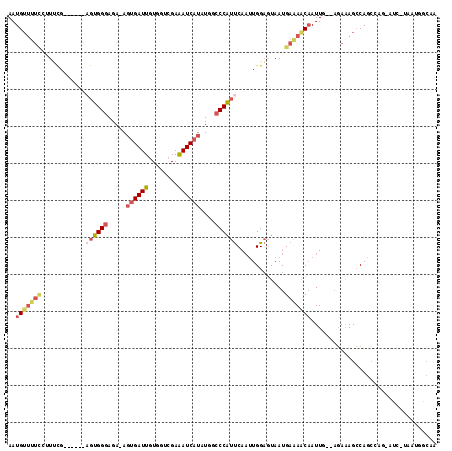
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:11 2006