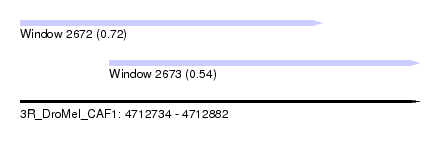
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,712,734 – 4,712,882 |
| Length | 148 |
| Max. P | 0.721204 |

| Location | 4,712,734 – 4,712,846 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.92 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
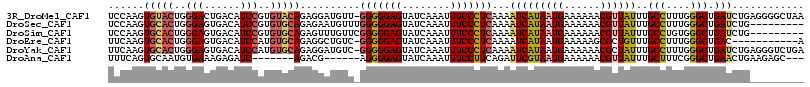
>3R_DroMel_CAF1 4712734 112 + 27905053 UAAAUAAUCUUCAACGAUA---GGAUAG---UAUUUUUUUCCAAGUGUACUGGGACUGACAUCCGUGUACAGAGGAUGUU-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA ......((((((.......---(((.((---.....)).)))...(((((..(((......)))..))))))))))).((-(((((((........))))))))).............. ( -28.30) >DroSec_CAF1 57967 114 + 1 UAAUUAAUCUUCAACGAUAGUUAGAUAG---U--UUUUUUCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGAAUGUUUGGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA ......((((..(((....)))))))..---.--..(((((....(((((..(((......)))..)))))..((...((((((((((........)))))))))).)).....))))) ( -27.40) >DroSim_CAF1 61250 111 + 1 UAAUUAAUCUUCAAGAAUA---CGAUAG---U--UUUUUUCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGUUUGUUCGGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA .........((((.(((((---.((...---.--.....))....(((((..(((......)))..))))).....)))))(((((((........)))))))..........)))).. ( -24.40) >DroEre_CAF1 61578 110 + 1 UAUUUUUACUACAAAUGAA---CUAUAG---A--GUUUAUUCAAGUGCACUGGGAGUGACAUCCAUGUGCAGAGGCUGUC-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA ...............((((---((....---)--)))))((((..(((((..(((......)))..))))).........-(((((((........)))))))..........)))).. ( -26.00) >DroYak_CAF1 60503 108 + 1 --UUUUUUCUUCAACAGAA---CGAUCG---A--AUUUUUUCAAGUGCACUGGGAGUGACAUCCAUGUGCAGAGGAUGUC-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA --.....(((((.......---.....(---(--(....)))...(((((..(((......)))..))))))))))....-(((((((........)))))))................ ( -27.00) >DroAna_CAF1 58964 96 + 1 UAGAA-------AACCAUA---UGAUAGCCAUACUUUACUUUCAGUGCAAUGUGAAAGAGAUC-------AGACG------AGGGGAGUAUCAAAUUUCCUUCAGAUUCGUAAUGAAAA .....-------...((((---(((.............((((((........)))))).....-------....(------((((((((.....)))))))))....)))).))).... ( -17.70) >consensus UAAUUAAUCUUCAACAAUA___CGAUAG___U__UUUUUUCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGGAUGUU_GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAA .............................................(((((..(((......)))..)))))..........(((((((........)))))))................ (-16.01 = -16.65 + 0.64)

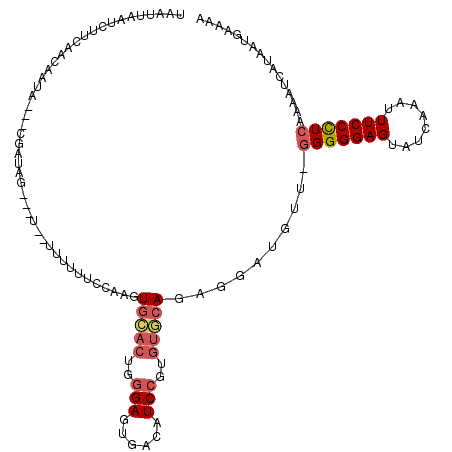
| Location | 4,712,767 – 4,712,882 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4712767 115 + 27905053 UCCAAGUGUACUGGGACUGACAUCCGUGUACAGAGGAUGUU-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAACGUUAUUUGCCUUUGGGCUGAUCUGAGGGGCUAA ....((((((((....(..((((((.........)))))).-.)...))))).......((((((..(((((((((......)))))...(((....))))))).))))))))).. ( -33.20) >DroSec_CAF1 58001 107 + 1 UCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGAAUGUUUGGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAACGUUAUUUGCCUUUGGGCUGAUCUG--------- ......(((((..(((......)))..))))).(((...((((((((((........)))))))))).((((((((......)))))...(((....))))))))).--------- ( -30.40) >DroSim_CAF1 61281 107 + 1 UCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGUUUGUUCGGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAACGUUAUUUGCCUGUGGGCUGAUCUG--------- ......(((((..(((......)))..)))))((.((((...((((((((.....)))))))))))).))((((((......))))))..(((....))).......--------- ( -27.50) >DroEre_CAF1 61609 104 + 1 UUCAAGUGCACUGGGAGUGACAUCCAUGUGCAGAGGCUGUC-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAGCGCUGUUUGCCUUUGGGCUGAC-----------A ......(((((..(((......)))..))))).(((((...-(((((((........)))))))....((.....))...))).))(((.(((....))).)))-----------. ( -30.90) >DroYak_CAF1 60532 115 + 1 UUCAAGUGCACUGGGAGUGACAUCCAUGUGCAGAGGAUGUC-GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAACGCUAUUUGCCUUUGGGCUGAUCUGAGGGUCUGA ((((..(((((..(((......)))..)))))...(((...-(((((((........)))))))...)))....))))............(((((..((....))..))))).... ( -32.20) >DroAna_CAF1 58993 100 + 1 UUUCAGUGCAAUGUGAAAGAGAUC-------AGACG------AGGGGAGUAUCAAAUUUCCUUCAGAUUCGUAAUGAAAAAACGUUAUUUGCUUUCGGGCUGAACUGAAGAGC--- (((((((.((.(.((((((.....-------....(------((((((((.....)))))))))......((((((......))))))...)))))).).)).)))))))...--- ( -26.80) >consensus UCCAAGUGCACUGGGAGUGACAUCCGUGUGCAGAGGAUGUU_GGGGGAGUAUCAAAUUUCCCUCAAAAUCAUAAUGAAAAAACGUUAUUUGCCUUUGGGCUGAUCUG_________ ......(((((..(((......)))..)))))..........(((((((........)))))))...(((((((((......))))))..(((....))).)))............ (-21.15 = -21.65 + 0.50)
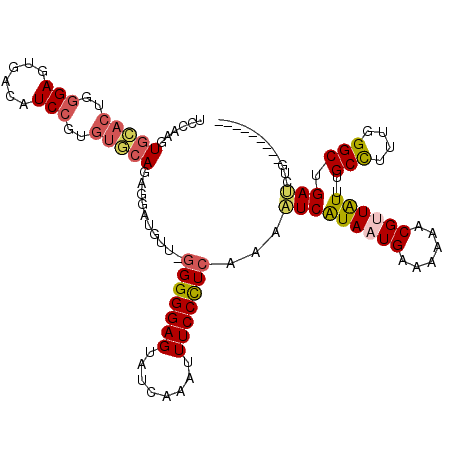
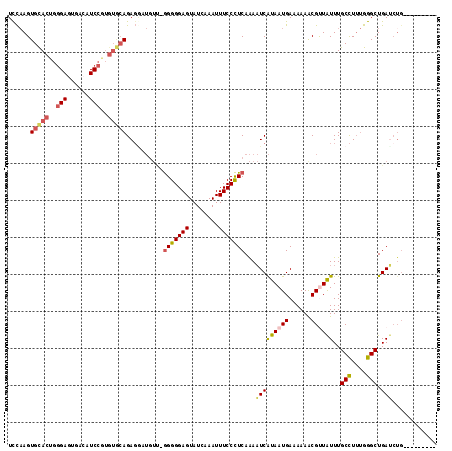
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:06 2006