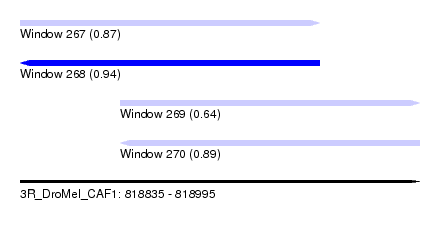
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 818,835 – 818,995 |
| Length | 160 |
| Max. P | 0.939511 |

| Location | 818,835 – 818,955 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 818835 120 + 27905053 AGGUUAAUAAUGCAAAGUAAUAGUACAGUAAUUUAUAACUUCAGUUUACAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCUGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGA ..........(((...(((....)))............((((.......(((((((((((......)))).(((....))))))))))((((((....)))))))))))))......... ( -25.60) >DroSim_CAF1 5991 105 + 1 GUUGUA---------------UGUAGAGUAAUUUAUAACUUCAGUUUUCAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCAGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGA (((((.---------------((((((....)))))).((((.......(((.(((((((......)))).(((....)))))).)))((((((....)))))))))))))))....... ( -23.30) >DroYak_CAF1 9296 109 + 1 A-----AAGCUUCAAAGUAAUAGUGCAGUAAUUUAUAACUUUAGUUUGUAUCAGGCUGGUGAUUACGCU------UCAUGUGCCUGAUAAUUACAAAAGUUACUGAAGGCAAUAAUAGGA .-----...(((((..(((....))).........(((((((.((...((((((((..((((.......------))))..))))))))...)).))))))).)))))............ ( -28.30) >consensus A___UAA___U_CAAAGUAAUAGUACAGUAAUUUAUAACUUCAGUUUACAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCUGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGA ................................((((..(((((((....(((((((((((......)))).((......)))))))))...(((....))))))))))...))))..... (-15.11 = -15.90 + 0.79)

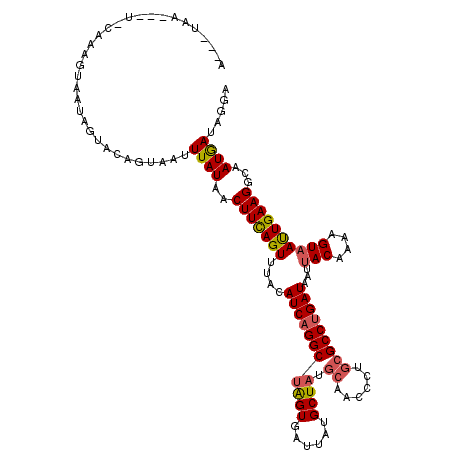
| Location | 818,835 – 818,955 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -15.21 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 818835 120 - 27905053 UCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCAGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGUAAACUGAAGUUAUAAAUUACUGUACUAUUACUUUGCAUUAUUAACCU ..........((((..((((((....))))))...))))...(((((((.(((........))))).(((((((((.((((((.(((......)))))).))).)))))))))..))))) ( -23.10) >DroSim_CAF1 5991 105 - 1 UCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCUGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGAAAACUGAAGUUAUAAAUUACUCUACA---------------UACAAC ............(((((.((((....))))(((((.(((..(((((((.......))))).)).))).)))))....))))).................---------------...... ( -17.30) >DroYak_CAF1 9296 109 - 1 UCCUAUUAUUGCCUUCAGUAACUUUUGUAAUUAUCAGGCACAUGA------AGCGUAAUCACCAGCCUGAUACAAACUAAAGUUAUAAAUUACUGCACUAUUACUUUGAAGCUU-----U ..........((.((((((((((((.((...((((((((...(((------.......)))...))))))))...)).))))))))....................))))))..-----. ( -20.95) >consensus UCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCAGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGAAAACUGAAGUUAUAAAUUACUGUACUAUUACUUUG_A___UUA___U .................((.(((((.((...((((((((....(((((.......)))))....))))))))...)).))))).)).................................. (-15.21 = -15.67 + 0.45)

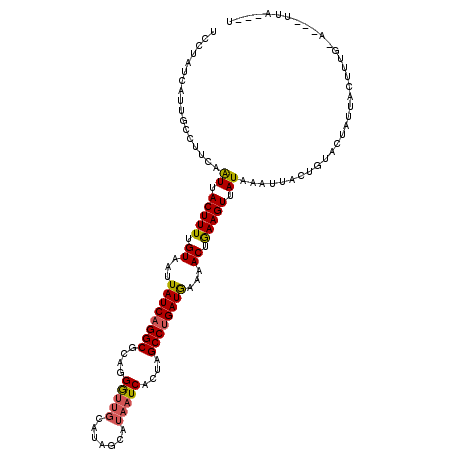
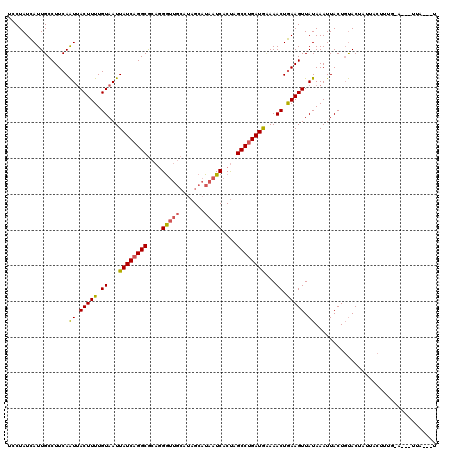
| Location | 818,875 – 818,995 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -21.94 |
| Energy contribution | -24.50 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 818875 120 + 27905053 UCAGUUUACAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCUGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGAGGCAGUUCCAUCCUGUGCCUCGUUAUUAUUUUGUAAAAGC ...((((..(((((((((((......)))).(((....))))))))))..(((((((((((((.((.((((..(((.((((....)))))))...)))))))))))..)))))))))))) ( -34.40) >DroSim_CAF1 6016 120 + 1 UCAGUUUUCAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCAGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGAGGCAGUUCCAUCCUGUGCCUCGUUAUUAUUUUGUAAAAGC ...(((((((((.(((((((......)))).(((....)))))).)))((((((....))))))))))))((((((((((((((...........)))))).)))))))).......... ( -31.00) >DroYak_CAF1 9331 112 + 1 UUAGUUUGUAUCAGGCUGGUGAUUACGCU------UCAUGUGCCUGAUAAUUACAAAAGUUACUGAAGGCAAUAAUAGGAGGCAGUUCCAAGCUGU--CUUGUUAUUAUUUUGUAAAAGG ........((((((((..((((.......------))))..)))))))).(((((((((((......)))((((((..((((((((.....)))))--))))))))).)))))))).... ( -33.00) >consensus UCAGUUUACAUCAGGCUAGUGAUUAUGCUAUGCAACCCUGCGCCUGAUAAUUACAAAAGUAAUUGAAGGCAAUGAUAGGAGGCAGUUCCAUCCUGUGCCUCGUUAUUAUUUUGUAAAAGC .........(((((((((((......)))).((......)))))))))..(((((((((((((...(((((..(((.((((....)))))))...))))).)))))..)))))))).... (-21.94 = -24.50 + 2.56)

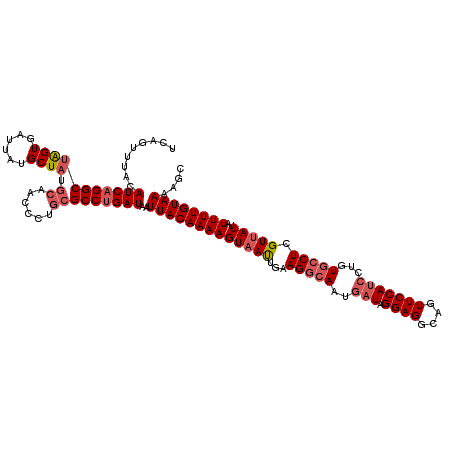
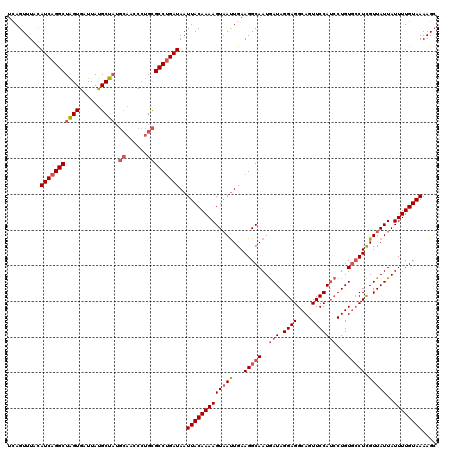
| Location | 818,875 – 818,995 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -23.41 |
| Energy contribution | -25.53 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 818875 120 - 27905053 GCUUUUACAAAAUAAUAACGAGGCACAGGAUGGAACUGCCUCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCAGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGUAAACUGA ....((((((((((((...((((((...((((((......))).)))..))))))..)))).)))))))).((((((((..(((((((.......))))).)).))))))))........ ( -34.20) >DroSim_CAF1 6016 120 - 1 GCUUUUACAAAAUAAUAACGAGGCACAGGAUGGAACUGCCUCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCUGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGAAAACUGA ....((((((((((((...((((((...((((((......))).)))..))))))..)))).))))))))(((((.(((..(((((((.......))))).)).))).)))))....... ( -31.20) >DroYak_CAF1 9331 112 - 1 CCUUUUACAAAAUAAUAACAAG--ACAGCUUGGAACUGCCUCCUAUUAUUGCCUUCAGUAACUUUUGUAAUUAUCAGGCACAUGA------AGCGUAAUCACCAGCCUGAUACAAACUAA ....((((((((......((((--....)))).............((((((....)))))).)))))))).((((((((...(((------.......)))...))))))))........ ( -21.30) >consensus GCUUUUACAAAAUAAUAACGAGGCACAGGAUGGAACUGCCUCCUAUCAUUGCCUUCAAUUACUUUUGUAAUUAUCAGGCGCAGGGUUGCAUAGCAUAAUCACUAGCCUGAUGAAAACUGA ....((((((((((((...((((((...((((((......))).)))..))))))..)))).)))))))).((((((((....(((((.......)))))....))))))))........ (-23.41 = -25.53 + 2.12)

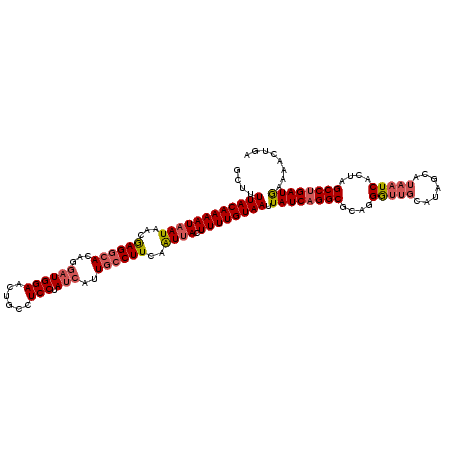
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:25 2006