
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,658,052 – 4,658,160 |
| Length | 108 |
| Max. P | 0.552861 |

| Location | 4,658,052 – 4,658,160 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
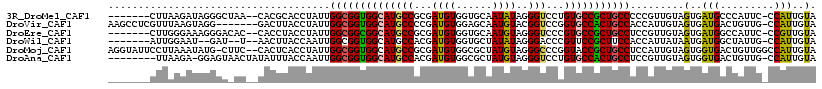
>3R_DroMel_CAF1 4658052 108 + 27905053 -------CUUAAGAUAGGGCUAA--CACGCACCUAUUGGCGGUGGCAUGCCGCGAUGUGGUGCAAUAUAGGGUCCUGUGCCGCUGCCCCCGUUGUAGUGAUGCCCAUUC-CCAUUGUA -------.........((((...--((((((.(....(((((((((..(((((...))))).....((((....)))))))))))))...).))).)))..))))....-........ ( -37.50) >DroVir_CAF1 9338 110 + 1 AAGCCUCGUUUAAGUAGG-------GACUUACCUAUUGGCGGUGGCAUGCCCCGAUGUGGAGCAAUGUACGGUCCGGUGCCACUGCCACCAUUGUAGUGAUGACUGUUG-CCAUUGUA ............((((((-------......))))))((((((((((.((.(((.((((.(....).))))...))).))...)))))))....((((....))))..)-))...... ( -35.70) >DroEre_CAF1 8034 108 + 1 -------CUUGGGAAAGGGACAC--CACCUACCUAUUGGCGGCGGCAUGCCGCGAUGUGGUGCAAUGUAGGGUCCCGUGCCGCUGCCUCCGUUGUAGUGAUGGCCAUUC-CCGUUGUA -------...(((((.((..((.--(((.(((.....((((((((((((((((...)))))........((...)))))))))))))......)))))).)).)).)))-))...... ( -43.70) >DroWil_CAF1 11077 104 + 1 -------AUUGGAAU--GAU--U--AACUUACCAAUUGGCGGUGGCAUGCCACGAUGUGGUGCUAUAUAGGGACCCGUUCCGCUUCCACCAUUAUAAUGAUGGCUAUUG-CCAUUGUA -------(((((...--...--.--......)))))..(((((((((.((((((..((((((.......((((....)))).....)))))).....)).))))...))-))))))). ( -31.32) >DroMoj_CAF1 9033 115 + 1 AGGUAUUCCUUAAAUAUG-CUUC--CACUCACCUAUUGGCGGUGGCAUGCCGCGAUGUGGCGCUAUGUAGGGCCCGGUACCGCUGCCUCCAUUGUAGUGGUGACUGUUGGCCAUUGUA (((....)))...(((((-(..(--(((.(.((....)).)))))...(((((...))))))).))))..((((((((((((((((.......)))))))).))))..))))...... ( -39.80) >DroAna_CAF1 11428 108 + 1 --------UUAAGA-GGAGUAACUAUAUUUACCAAUUGGCGGUGGCAUGCCACGAUGUGGCGCUAUGUAGGGUCCUGUGCCACUGCCUCCGUUGUAGUGGUGACUGUUG-CCAUUGUA --------......-(((((((......)))).....((((((((((((((((...))))).......((....))))))))))))))))....((((((..(...)..-)))))).. ( -38.50) >consensus _______CUUAAAAUAGGGC_AC__CACUUACCUAUUGGCGGUGGCAUGCCGCGAUGUGGUGCAAUGUAGGGUCCCGUGCCGCUGCCUCCAUUGUAGUGAUGACUAUUG_CCAUUGUA .....................................((((((((((((((...((((......))))..)))...))))))))))).........(..(((.........)))..). (-23.72 = -23.20 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:50 2006