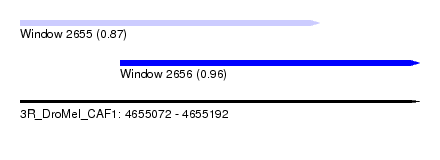
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,655,072 – 4,655,192 |
| Length | 120 |
| Max. P | 0.959560 |

| Location | 4,655,072 – 4,655,162 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -19.12 |
| Energy contribution | -18.65 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4655072 90 + 27905053 AUUGCU------CCACCAUUCCUUCAUUGUGCAGAUUAUUUUCAAUAGCUGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGA------UCACGCU ......------..................((.((......)).....(((((((..(((((((.......)))))))..)))))))..------....)). ( -21.60) >DroSec_CAF1 4803 90 + 1 CUUGCU------CCACCAUUCCCUCGUUGUGCAGAUUAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGA------UCAUUCU ......------.......((((..(((((...((......)).))))).(((((..(((((((.......)))))))..)))))))))------....... ( -25.20) >DroSim_CAF1 5019 90 + 1 CUUGUU------CCACCAUUCCCUCGUUGUGCAGAUUAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCAGCAGAGCUCUCAGCGGGA------UCAUUCU ......------.......((((..(((((...((......)).))))).(((((..((((((.((....))))))))..)))))))))------....... ( -25.80) >DroEre_CAF1 5089 90 + 1 UUUGCU------CCACCAUUCCCUCGUUGUGUAGAUUGUUUUCAAUGGCCGUUGGCAGCUCUGUCUGAUCCGCAGAGCUUUCAGCGGGA------UCAUCCC ......------.......((((((((((.............))))))..(((((.((((((((.......)))))))).)))))))))------....... ( -26.22) >DroYak_CAF1 5213 90 + 1 CUUGUU------CCACCAUUCCCUCGUUGUGUAGAUUAUUUUCAAUGGCCGUUGGAAGCUCUGUCUGAUCCACAGAGCUCUCAGCGGGA------UCAUUCU ......------.......((((((((((.............))))))..((((..((((((((.......))))))))..))))))))------....... ( -27.22) >DroAna_CAF1 8441 102 + 1 UUUCAUUGGCCAGCAACAUGGCCUCAUUGUGAUGGUUGUUUUCAAUGGCCGUGACCUGUUCUGUGGCCUCCAUGGAGCUCUCAUCGGGGAUUUCGUCACCCU .......(((((..((.(..(((((.....)).)))..).))...)))))(((((..(((((((((...)))))))))((((....))))....)))))... ( -33.70) >consensus CUUGCU______CCACCAUUCCCUCGUUGUGCAGAUUAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGA______UCAUUCU ............................(((..((......)).....(((((((..(((((((.......)))))))..))))))).........)))... (-19.12 = -18.65 + -0.47)


| Location | 4,655,102 – 4,655,192 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4655102 90 + 27905053 UAUUUUCAAUAGCUGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGAUCACGCUGAUCCUGAACCUCCUC---CUCUUCGUCA---UCAU ....(((.......(((((..(((((((.......)))))))..)))))((((((.....))))))))).......---..........---.... ( -24.90) >DroGri_CAF1 5926 93 + 1 GAUUCUCAAUUGUGGUCGGUUGCUCUGCUGGCUCUGCAGAGCUAUCUGCCUCCUGAUCCAGUUCCGGAUCCGCCUGCUGCUCGGCAUCG---UCAU (((..........(((.(((.(((((((.......))))))).))).)))....(((((......))))).(((........)))...)---)).. ( -30.30) >DroSec_CAF1 4833 90 + 1 UAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGAUCAUUCUGAUCCUGAACCUCUUC---CUCUUCGUCA---UCAU ....((((.....((((((..(((((((.......)))))))..))))))(((((.....))))))))).......---..........---.... ( -26.60) >DroSim_CAF1 5049 90 + 1 UAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCAGCAGAGCUCUCAGCGGGAUCAUUCUGAUCCUGAACCUCCUC---AUCUUCGUCA---UCAU ....((((.....((((((..((((((.((....))))))))..))))))(((((.....))))))))).......---..........---.... ( -27.20) >DroEre_CAF1 5119 90 + 1 UGUUUUCAAUGGCCGUUGGCAGCUCUGUCUGAUCCGCAGAGCUUUCAGCGGGAUCAUCCCGAGCCUGAGCCUGCUC---UUCUUCGUCG---UCAU ........(((((.(((((.((((((((.......)))))))).)))))(((.....)))((((........))))---.........)---)))) ( -30.70) >DroYak_CAF1 5243 93 + 1 UAUUUUCAAUGGCCGUUGGAAGCUCUGUCUGAUCCACAGAGCUCUCAGCGGGAUCAUUCUGAUCCUGAACCUCCUC---CUCUUGGUCGUCAUCAU ........((((((((((..((((((((.......))))))))..))))((((((.....))))))..........---.....))))))...... ( -34.00) >consensus UAUUUUCAAUAGCCGUUGGCUGCUCUGUCUGAUCCGCAGAGCUCUCAGCGGGAUCAUUCUGAUCCUGAACCUCCUC___CUCUUCGUCA___UCAU ....((((.....((((((..(((((((.......)))))))..))))))(((((.....)))))))))........................... (-21.46 = -22.18 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:49 2006