
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,575,771 – 4,575,931 |
| Length | 160 |
| Max. P | 0.799513 |

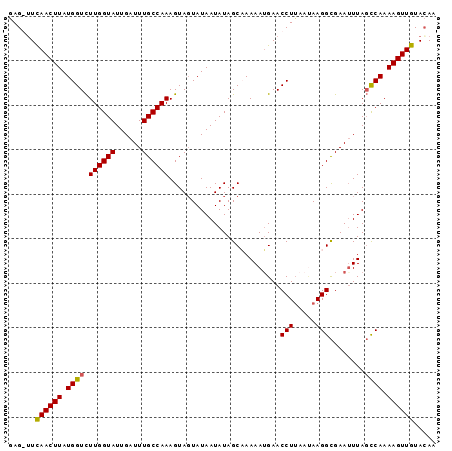
| Location | 4,575,771 – 4,575,862 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -15.88 |
| Energy contribution | -15.75 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4575771 91 + 27905053 GAG-GUUAACUUAUGGUCUUGGUAUUGAUUUGCCAAAGUAUUAUAAUAUAGCAAAAAUGAACCU----AAGGCGAAUUUAGCCAAAAGUUGUACAA ...-((((((((......(((((...((((((((..((..((((............))))..))----..))))))))..))))))))))).)).. ( -17.40) >DroSec_CAF1 32561 95 + 1 GAG-UUCAACUUAUGGUCUUGGUAUUGAUUUGCCAAAGUAGUAUAAUAUAGCAAAAAUGAACCUUAAUAAGGCGAAUUUAGCCAAAAGUUGUACAA ..(-(.((((((.((((.((((((......))))))..................((((...(((.....)))...)))).)))).)))))).)).. ( -20.50) >DroEre_CAF1 33441 96 + 1 AAGGUUCAACUUAUGGUCUUGGUAUUGAUUUGCCAAAGUAGUAUAAUAUAGCCAAAAGGAACCUUAAUAAGGCGAAUUUAGCCAAAAGUUGUAGAA (((((((..(((.((((.((((((......)))))).(((......))).)))).)))))))))).....(((.......)))............. ( -23.40) >DroYak_CAF1 33143 96 + 1 GAGGUUCAACUUAUGGUUUUGGUAUUGAUUUGCCAAAGUAGUAUAAUAUAGCCAAAAUGAACCUUAACAAGGUGAAAUUACUCAAAAGUUGUACAA ((((((((..((.(((((((((((......)))))))(((......))).)))).)))))))))).((((..(((......)))....)))).... ( -20.90) >consensus GAG_UUCAACUUAUGGUCUUGGUAUUGAUUUGCCAAAGUAGUAUAAUAUAGCAAAAAUGAACCUUAAUAAGGCGAAUUUAGCCAAAAGUUGUACAA ......((((((.((((.((((((......)))))).........................(((.....)))........)))).))))))..... (-15.88 = -15.75 + -0.13)


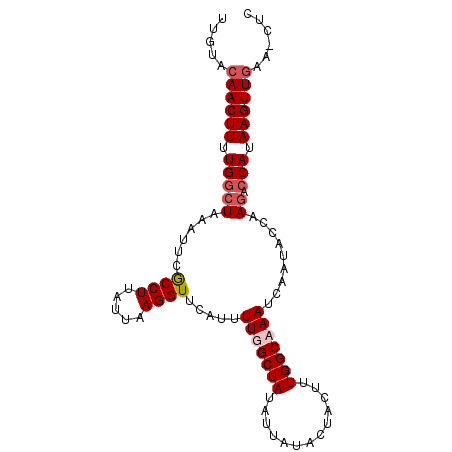
| Location | 4,575,771 – 4,575,862 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -16.36 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.92 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4575771 91 - 27905053 UUGUACAACUUUUGGCUAAAUUCGCCUU----AGGUUCAUUUUUGCUAUAUUAUAAUACUUUGGCAAAUCAAUACCAAGACCAUAAGUUAAC-CUC ......(((((..(((.......)))..----.(((.....(((((((.............))))))).....)))........)))))...-... ( -14.82) >DroSec_CAF1 32561 95 - 1 UUGUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCAUUUUUGCUAUAUUAUACUACUUUGGCAAAUCAAUACCAAGACCAUAAGUUGAA-CUC ..((.((((((.(((((......((((.....)))).....(((((((.............))))))).........)).))).)))))).)-).. ( -19.52) >DroEre_CAF1 33441 96 - 1 UUCUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCCUUUUGGCUAUAUUAUACUACUUUGGCAAAUCAAUACCAAGACCAUAAGUUGAACCUU .....((((((.((((((((...((((.....))))....)))))).............(((((..........)))))..)).))))))...... ( -18.00) >DroYak_CAF1 33143 96 - 1 UUGUACAACUUUUGAGUAAUUUCACCUUGUUAAGGUUCAUUUUGGCUAUAUUAUACUACUUUGGCAAAUCAAUACCAAAACCAUAAGUUGAACCUC ..((.((((((.((..((((........)))).(((........((((.............))))........))).....)).)))))).))... ( -13.11) >consensus UUGUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCAUUUUGGCUAUAUUAUACUACUUUGGCAAAUCAAUACCAAGACCAUAAGUUGAA_CUC .....((((((.(((((......((((.....)))).....(((((((.............))))))).........)).))).))))))...... (-12.86 = -13.92 + 1.06)

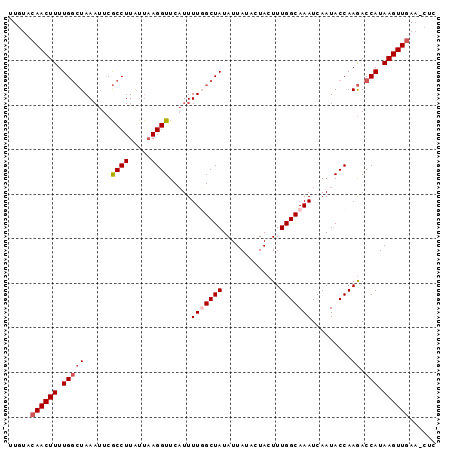
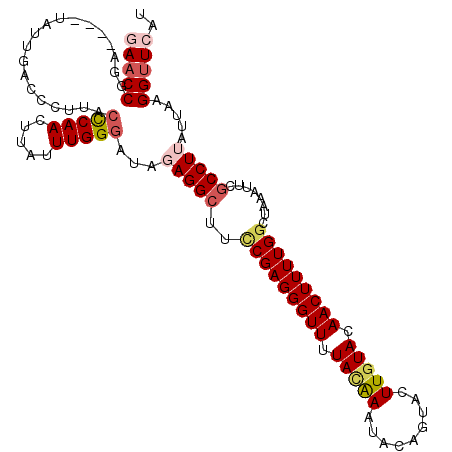
| Location | 4,575,826 – 4,575,931 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.46 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4575826 105 - 27905053 GAACCGGA----UAUUGACCCUUACCCAACUUAUUUGGGAUAGAGGCUUUCGAGGGUUUUACAAAUACAGUACUUGUACAACUUUUGGCUAAAUUCGCCUU----AGGUUCAU (((((((.----.......))...(((((.....)))))...(((((..(((((((((.(((((.........))))).)))))))))........)))))----.))))).. ( -27.70) >DroSec_CAF1 32616 113 - 1 AAACCGGAUGGAUAUUGACCCUUACCCAACUUAUUUGGGAUAGAGGCUUCCGAGGGUUUUACAAAUACAGUACUUGUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCAU ...((....))....(((.((((((((((.....)))))...(((((..(((((((((.(((((.........))))).)))))))))........)))))..))))).))). ( -29.40) >DroSim_CAF1 33155 113 - 1 AAACCGGAUGAAUAUUGACCCUUACCCAACUUAUUUGGGAUAGAGGCUUCCGAGGGUUUUACAAAUACAGUACUUGUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCAU ...............(((.((((((((((.....)))))...(((((..(((((((((.(((((.........))))).)))))))))........)))))..))))).))). ( -29.00) >DroEre_CAF1 33497 107 - 1 GAACCGGA----UACAAGCCCUUACUCAACUUAUUUGGUAC--AGGCUUCCGAGGGUUUUAUAAUUAUAAUACUUCUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCCU ((((((((----....((((..(((.(((.....)))))).--.)))))))((((...((((....))))..))))..........(((.......))).......))))).. ( -24.40) >DroYak_CAF1 33199 107 - 1 GAUCCGGA----UAGUAGCCCUUACUCAACUUAUUUGGGAU--AGGCUUCCGAGGGUUUUAUGAUUAUAGUACUUGUACAACUUUUGAGUAAUUUCACCUUGUUAAGGUUCAU ((.(((((----....((((..(.(((((.....)))))).--.)))))))((((..............(((....)))......((((....)))))))).....)).)).. ( -21.10) >consensus GAACCGGA____UAUUGACCCUUACCCAACUUAUUUGGGAUAGAGGCUUCCGAGGGUUUUACAAAUACAGUACUUGUACAACUUUUGGCUAAAUUCGCCUUAUUAAGGUUCAU (((((...................(((((.....)))))...(((((..(((((((((.(((((.........))))).)))))))))........))))).....))))).. (-19.30 = -20.46 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:30 2006