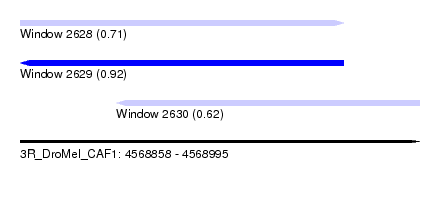
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,568,858 – 4,568,995 |
| Length | 137 |
| Max. P | 0.915082 |

| Location | 4,568,858 – 4,568,969 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.23 |
| Energy contribution | -28.85 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
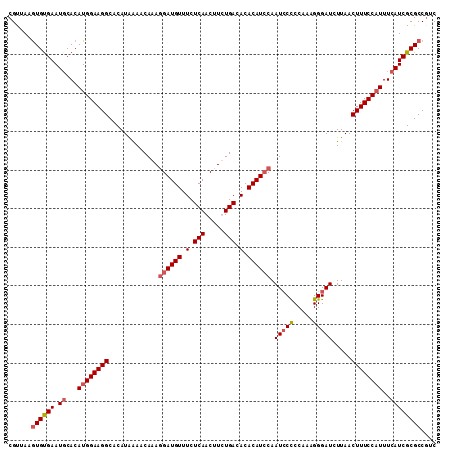
>3R_DroMel_CAF1 4568858 111 + 27905053 GAAGGCGCGAUGAAAUGGAAAGUUAAGAUCCCUUUGGAGGAUUGGAUGUGUGUCAGAAGUUGAGAAACAUCCUUUGUUUUAUGUGCCUUCCAUGUGCAUUCGCACUUAACG ((((((((.((((((..((.......((((((....).)))))((((((.(.((((...)))).).))))))))..))))))))))))))...((((....))))...... ( -38.90) >DroSec_CAF1 25595 111 + 1 GACGGCGCGAUAAAAUGGAAAGUUAAGAUCCCUUUGGGGGAUUGCAUGUGUGUCAGAAGUUGAGAAACAUCCUUUGUUCCAUGUGCCUUCCAUGUGAAUUCACAUUUAACG ((.(((((......((((((......((((((.....))))))(.((((.(.((((...)))).).)))).)....))))))))))).)).(((((....)))))...... ( -28.60) >DroEre_CAF1 26359 111 + 1 GCCGGCGCGAUGAAAAGGAAAGUUAAGAUCCCUUUGGGGGAUUGGAUGUGUGUCAAAAGUUGAGAAACAUCUUUUGUUUUAUGUGCCUUCCAUGUGCAUUCACACUUAGCG ((.(((((.((((((..(((......((((((.....)))))).(((((.(.(((.....))).).))))))))..))))))))))).....((((....))))....)). ( -31.20) >DroYak_CAF1 25993 111 + 1 GACGGCGCGAUGAAAUGGAAAGUUAAGAUCCCAUUGGGGCAUUGGAUGUGUGUCAAAAGUUGAGAAACAUCCUUUGUUUUAUGUGCCUUCCAUGUGCAUUCACACUUAACG ((.(((((.((((((..((.......((((((....))).)))((((((.(.(((.....))).).))))))))..))))))))))).))..((((....))))....... ( -31.50) >consensus GACGGCGCGAUGAAAUGGAAAGUUAAGAUCCCUUUGGGGGAUUGGAUGUGUGUCAAAAGUUGAGAAACAUCCUUUGUUUUAUGUGCCUUCCAUGUGCAUUCACACUUAACG ...(((((.((((((..((.......((((((.....))))))((((((.(.((((...)))).).))))))))..))))))))))).....((((....))))....... (-28.23 = -28.85 + 0.62)

| Location | 4,568,858 – 4,568,969 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.80 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
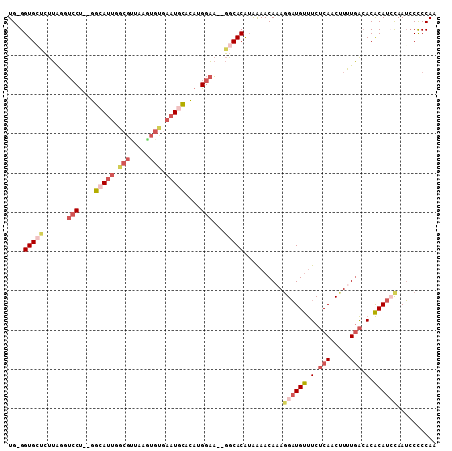
>3R_DroMel_CAF1 4568858 111 - 27905053 CGUUAAGUGCGAAUGCACAUGGAAGGCACAUAAAACAAAGGAUGUUUCUCAACUUCUGACACACAUCCAAUCCUCCAAAGGGAUCUUAACUUUCCAUUUCAUCGCGCCUUC ......((((((.((...((((((((.............((((((.(.(((.....))).).)))))).(((((.....))))).....))))))))..)))))))).... ( -30.70) >DroSec_CAF1 25595 111 - 1 CGUUAAAUGUGAAUUCACAUGGAAGGCACAUGGAACAAAGGAUGUUUCUCAACUUCUGACACACAUGCAAUCCCCCAAAGGGAUCUUAACUUUCCAUUUUAUCGCGCCGUC ..((..(((((....)))))..))(((.(((((((....(.((((.(.(((.....))).).)))).).(((((.....))))).......))))))......).)))... ( -23.90) >DroEre_CAF1 26359 111 - 1 CGCUAAGUGUGAAUGCACAUGGAAGGCACAUAAAACAAAAGAUGUUUCUCAACUUUUGACACACAUCCAAUCCCCCAAAGGGAUCUUAACUUUCCUUUUCAUCGCGCCGGC .(((..((((((.((.....((((((.........((((((.((.....)).))))))...........(((((.....))))).....))))))....)))))))).))) ( -26.50) >DroYak_CAF1 25993 111 - 1 CGUUAAGUGUGAAUGCACAUGGAAGGCACAUAAAACAAAGGAUGUUUCUCAACUUUUGACACACAUCCAAUGCCCCAAUGGGAUCUUAACUUUCCAUUUCAUCGCGCCGUC ......((((((.((...((((((((.............((((((.(.(((.....))).).))))))....(((....))).......))))))))..)))))))).... ( -28.30) >consensus CGUUAAGUGUGAAUGCACAUGGAAGGCACAUAAAACAAAGGAUGUUUCUCAACUUCUGACACACAUCCAAUCCCCCAAAGGGAUCUUAACUUUCCAUUUCAUCGCGCCGUC ......((((((.((...((((((((.............((((((.(.(((.....))).).)))))).(((((.....))))).....))))))))..)))))))).... (-22.67 = -23.80 + 1.13)

| Location | 4,568,891 – 4,568,995 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -13.58 |
| Energy contribution | -16.22 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
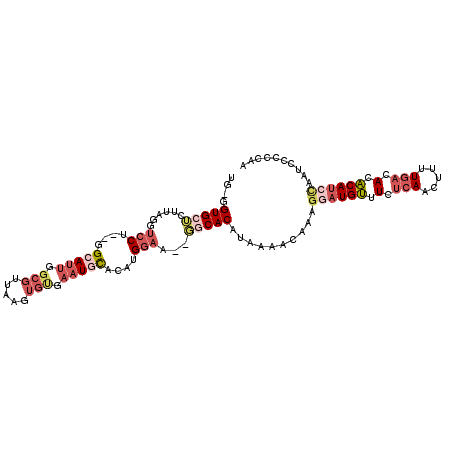
>3R_DroMel_CAF1 4568891 104 - 27905053 UG-GGUGCUCUUAGGUCCU--GGCAUUGGCGUUAAGUGCGAAUGCACAUGGAA--GGCACAUAAAACAAAGGAUGUUUCUCAACUUCUGACACACAUCCAAUCCUCCAA ((-((((((......((((--((((((.(((.....))).))))).)).))).--)))))..........((((((.(.(((.....))).).))))))......))). ( -31.70) >DroSec_CAF1 25628 106 - 1 UG-GGUGCUCUUAGCUCCUUUGGCAUUGGCGUUAAAUGUGAAUUCACAUGGAA--GGCACAUGGAACAAAGGAUGUUUCUCAACUUCUGACACACAUGCAAUCCCCCAA ((-(((((.......(((((((....((((.((..(((((....)))))..))--.)).)).....)))))))(((.((.........)).)))...)))....)))). ( -28.20) >DroEre_CAF1 26392 104 - 1 UG-GGUGGUCUUAGGUCCU--GGCAUUGGCGCUAAGUGUGAAUGCACAUGGAA--GGCACAUAAAACAAAAGAUGUUUCUCAACUUUUGACACACAUCCAAUCCCCCAA ((-((((((((....((((--((((((.((((...)))).))))).)).))))--)))........((((((.((.....)).)))))).....))))))......... ( -30.70) >DroYak_CAF1 26026 105 - 1 UGGGGUGGACUUAGGUCCU--GGCAUUGGCGUUAAGUGUGAAUGCACAUGGAA--GGCACAUAAAACAAAGGAUGUUUCUCAACUUUUGACACACAUCCAAUGCCCCAA ((((((((((....))))(--(....((((.((..(((((....)))))..))--.)).)).....))..((((((.(.(((.....))).).))))))...)))))). ( -34.70) >DroAna_CAF1 21584 94 - 1 UG-GGUGCGGCUGGGGGCC--GGAAGGAGAGUU---------UCUAUUUGGGUCCUGCACG-AAAACAAAGGCUGCCUCACA-CAUUUAGGACGCAU-UAAGCCUCCAA ((-((.((((((.((((((--.(((..((....---------.)).))).))))))....(-....)...))))))))))..-......(((.((..-...)).))).. ( -30.00) >consensus UG_GGUGCUCUUAGGUCCU__GGCAUUGGCGUUAAGUGUGAAUGCACAUGGAA__GGCACAUAAAACAAAGGAUGUUUCUCAACUUUUGACACACAUCCAAUCCCCCAA ....(((((......(((....(((((.(((.....))).)))))....)))...)))))..........((((((.(.(((.....))).).)))))).......... (-13.58 = -16.22 + 2.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:24 2006