
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,566,166 – 4,566,360 |
| Length | 194 |
| Max. P | 0.950038 |

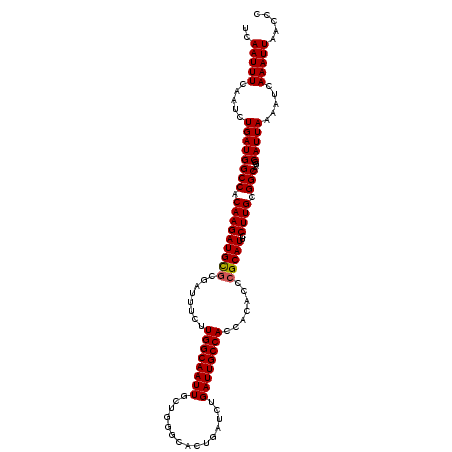
| Location | 4,566,166 – 4,566,281 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4566166 115 + 27905053 UCAAUUUCAAUCUGAUGGCCCCAAGAUGCGCGAUUUCUUGGCAAUUGCUGGGCACUGAUCCGAUUGCCACCACACCCGCAUUCCUUGCGGCAAAGCAUUAAAAUCAAAUUAACCC .......(((((.(((.(((((((((.........)))))((....)).))))....))).))))).........(((((.....)))))......................... ( -25.40) >DroSec_CAF1 23025 111 + 1 UCAAUUU----CUGAUGGCCACAAGAUGCGCGAUUUCUUGGCAAUUGCUGUGCACUGAUCUGAUUGCCACCACACCCGCAUUCCUUGCGGCAAGGCAUUAAAAUGAAAUUAACCC ..(((((----(((((((((.(((((((((.(......((((((((...............))))))))......))))))..)))).)))....)))))....))))))..... ( -26.46) >DroSim_CAF1 23733 111 + 1 UCAAUUU----CUGAUGGCCACAAGAUGCACGAUUUCUUGGCAAUUGCUGUGCACUGAUCUGAUUGCCACCACACCCGCAUUCCUUGCGGCAGGGCAUUAAAAUCAAAUUAACCC .....((----(((.((((....(((((((((....)..(((....))))))))....)))....))))......(((((.....)))))))))).................... ( -24.60) >DroEre_CAF1 23538 115 + 1 UGAAUUUCAAUCUGAUGGCCACAAGAUGUGCGAUUUCUUGGCAAUUGCUGGGCACUGAUCUGAUUGCCACCACACCCGCAUUCCUUGUGGCAAAGCAUUAAAAUCAAAUUAACCC (((.(((.(((((....((((((((..(((((......((((((((...(((......))))))))))).......)))))..))))))))..)).)))))).)))......... ( -32.32) >DroYak_CAF1 23281 115 + 1 UCAAUUUCAAUCUGAUGGCCACAAGAUGCGCGAUUUCUUGGCAAUUGUUGGGCACUGAUCUGAUUGCCACCACACCCGCAUUCCUUGUGGCAAGGCAUUAAAAUCAAAUUAACCC ............((((((((((((((((((.(......((((((((((..(...)..))..))))))))......))))))..))))))))....)))))............... ( -29.80) >consensus UCAAUUUCAAUCUGAUGGCCACAAGAUGCGCGAUUUCUUGGCAAUUGCUGGGCACUGAUCUGAUUGCCACCACACCCGCAUUCCUUGCGGCAAGGCAUUAAAAUCAAAUUAACCC ..(((((.....((((((((.(((((((((........((((((((...............)))))))).......)))))..)))).)))....))))).....)))))..... (-22.84 = -22.88 + 0.04)

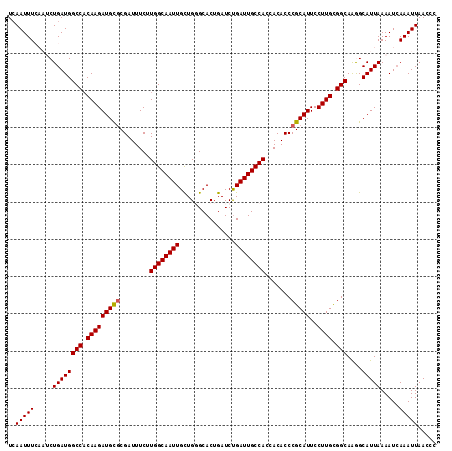
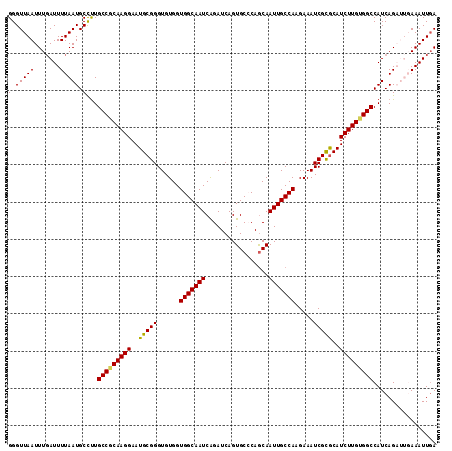
| Location | 4,566,166 – 4,566,281 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -28.87 |
| Energy contribution | -28.59 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4566166 115 - 27905053 GGGUUAAUUUGAUUUUAAUGCUUUGCCGCAAGGAAUGCGGGUGUGGUGGCAAUCGGAUCAGUGCCCAGCAAUUGCCAAGAAAUCGCGCAUCUUGGGGCCAUCAGAUUGAAAUUGA ........(..((((((((.((..(((.((((..((((((((....(((((((.((........))....)))))))....))).))))))))).)))....))))))))))..) ( -35.00) >DroSec_CAF1 23025 111 - 1 GGGUUAAUUUCAUUUUAAUGCCUUGCCGCAAGGAAUGCGGGUGUGGUGGCAAUCAGAUCAGUGCACAGCAAUUGCCAAGAAAUCGCGCAUCUUGUGGCCAUCAG----AAAUUGA ...((((((((......((((((..((....)).....))))))((((((.((.((((..((((...((....))...(....))))))))).)).)))))).)----))))))) ( -32.60) >DroSim_CAF1 23733 111 - 1 GGGUUAAUUUGAUUUUAAUGCCCUGCCGCAAGGAAUGCGGGUGUGGUGGCAAUCAGAUCAGUGCACAGCAAUUGCCAAGAAAUCGUGCAUCUUGUGGCCAUCAG----AAAUUGA ........(..(((((.((((((..((....)).....))))))((((((...(((....((((((.((....))...(....))))))).)))..)))))).)----))))..) ( -32.20) >DroEre_CAF1 23538 115 - 1 GGGUUAAUUUGAUUUUAAUGCUUUGCCACAAGGAAUGCGGGUGUGGUGGCAAUCAGAUCAGUGCCCAGCAAUUGCCAAGAAAUCGCACAUCUUGUGGCCAUCAGAUUGAAAUUCA ...((((((((((...........(((((((((..(((((......(((((((........(((...)))))))))).....)))))..))))))))).))))))))))...... ( -35.10) >DroYak_CAF1 23281 115 - 1 GGGUUAAUUUGAUUUUAAUGCCUUGCCACAAGGAAUGCGGGUGUGGUGGCAAUCAGAUCAGUGCCCAACAAUUGCCAAGAAAUCGCGCAUCUUGUGGCCAUCAGAUUGAAAUUGA ........(..((((((((.....((((((((..((((((((....(((((((.................)))))))....))).)))))))))))))......))))))))..) ( -34.73) >consensus GGGUUAAUUUGAUUUUAAUGCCUUGCCGCAAGGAAUGCGGGUGUGGUGGCAAUCAGAUCAGUGCCCAGCAAUUGCCAAGAAAUCGCGCAUCUUGUGGCCAUCAGAUUGAAAUUGA (.(((((.......))))).)...(((((((((..(((((......(((((((.................))))))).....)))))..)))))))))................. (-28.87 = -28.59 + -0.28)


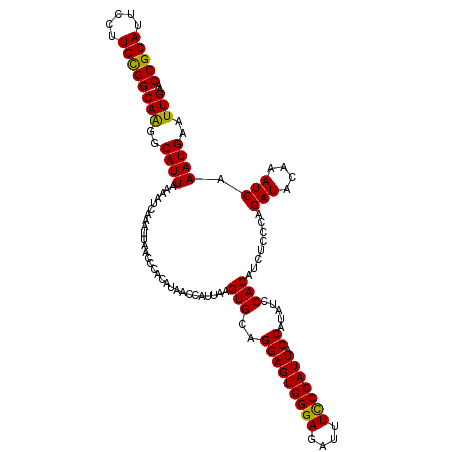
| Location | 4,566,241 – 4,566,360 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.55 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -22.10 |
| Energy contribution | -21.46 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.30 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4566241 119 + 27905053 CCGCAUUCCUUGCGGCAAAGCAUUAAAAUCAAAUUAACCCACAUAACCAUUAAGUGCAGGAGUGGGAGAUUUCCCAUUACCAUAUCCACAUUUCCCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))(((..((((.............................(((..((((((((......)))))).)).....)))........(((....))).))))..))).. ( -22.80) >DroSec_CAF1 23096 119 + 1 CCGCAUUCCUUGCGGCAAGGCAUUAAAAUGAAAUUAACCCAUAUAACCAUUAAGUGCAGGAGUGGGAGCUUUUCCAUUACCAUAUCCACAUCUCUCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))(((..((((.............................(((..(((((((((....))))))).)).....)))........(((....))).))))..))).. ( -21.00) >DroSim_CAF1 23804 119 + 1 CCGCAUUCCUUGCGGCAGGGCAUUAAAAUCAAAUUAACCCACAUAACCAUUAAGUGCAGGAGUGGGAGAUUUUCCAUUACCAUAUCCACAUAUCCCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))..(((...................(((...........)))..(((((((((....))))))).))............))).....(((.((....)).))).. ( -21.10) >DroEre_CAF1 23613 119 + 1 CCGCAUUCCUUGUGGCAAAGCAUUAAAAUCAAAUUAACCCACAUAACCAUUACGUGCAGGAGUGGGAGAUUUCCCAUUACCAUAUCCACAUCUCCCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))(((..((((.............................(((..((((((((......)))))).)).....)))........(((....))).))))..))).. ( -21.00) >DroYak_CAF1 23356 119 + 1 CCGCAUUCCUUGUGGCAAGGCAUUAAAAUCAAAUUAACCCACAUAAUCAUUAAGUGCAGGAGUGGGAGAUUUCCCAUUACCAUAUCCACAUCUCCCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))(((..((((.............................(((..((((((((......)))))).)).....)))........(((....))).))))..))).. ( -22.10) >consensus CCGCAUUCCUUGCGGCAAGGCAUUAAAAUCAAAUUAACCCACAUAACCAUUAAGUGCAGGAGUGGGAGAUUUCCCAUUACCAUAUCCACAUCUCCCAGAUACAAAUCAAAUGAAUUGAA (((((.....)))))(((..((((.............................(((..(((((((((....))))))).)).....)))........(((....))).))))..))).. (-22.10 = -21.46 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:22 2006