| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,561,263 – 4,561,374 |
| Length | 111 |
| Max. P | 0.814534 |

| Location | 4,561,263 – 4,561,374 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
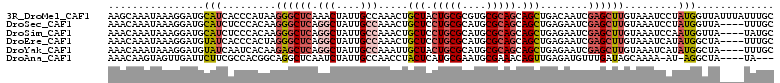
>3R_DroMel_CAF1 4561263 111 + 27905053 AAGCAAAUAAAGGAUGCAUCACCCAUAAGGGCUCAAACUAUUGCCAAACUGCUACUGCGCGUGCGCAGCAGCUGACAAUCGAGCUUGUAAAUCCUAUGGUUAUUUAUUUGC ..(((((((((.(((.(((.........((((((.....((((.((....(((.(((((....))))).))))).)))).)))))).........)))))).))))))))) ( -28.87) >DroSec_CAF1 18138 107 + 1 AAACAAAUAAAGGAUGCAUCUCCCACAAGGGCUCAGGCUAUUGCCAAACUGCUCCUGCGCAUGCGCAGCAGCUGAGAAUCGAGCUUGUAAAUCCUAUGGUUA----UUUGC ...(((((((((((((((.(((((....)).(((((((....))).....(((.(((((....))))).)))))))....)))..)))..)))))....)))----)))). ( -34.00) >DroSim_CAF1 18838 107 + 1 AAACAAAUAAAGGAUGCAUCUCCCACAAGGGCUCAGGCUAUUGCCAAACUGCUCCUGCGCAUGCGCAGCAGCUGAGAAUCGAGCUUGUAAAUCCAAUGGUUA----UAUGC ...........(((((((.(((((....)).(((((((....))).....(((.(((((....))))).)))))))....)))..)))..))))........----..... ( -30.30) >DroEre_CAF1 18579 107 + 1 AAACAAAUAAAGGAUGUAUCACCCACUAGGGCUCAGGCUAUUGCCAAACUGCUCCUGCGCAUGCGCAGCAGCUGAGAAUCGAGCUUGUAAAUCAUAUGGCUA----UUUGC ...((((((...(((......(((....)))(((((((....))).....(((.(((((....))))).))))))).)))..(((............)))))----)))). ( -29.60) >DroYak_CAF1 18373 107 + 1 AAACAAAUAAAGGAUGUAUCAAUCACAAGAGCUCAGGCUAUUGCCAAAUUGCUACUGCGCAUGCGCAGCAGCUGAGAAUCGAGCUUGUAAAUCAUAUGGCUA----UUUGC ...((((((...(((......)))....((((((.(((....))).....(((.(((((....))))).)))........))))))..............))----)))). ( -30.80) >DroAna_CAF1 15321 102 + 1 AAACAAGUAGUUGAUUCUUCGCCACGGCAGGCUCAAUCUAUUGCCAACCUACUCAUGCGAAUGCGAAACAGUUGAGAUGUUUGAUAGCAAAA-AU-AGGCUA----UA--- (((((..((..((.((((((((...((..(((..........)))..)).......)))))...))).))..))...))))).(((((....-..-..))))----).--- ( -18.70) >consensus AAACAAAUAAAGGAUGCAUCACCCACAAGGGCUCAGGCUAUUGCCAAACUGCUCCUGCGCAUGCGCAGCAGCUGAGAAUCGAGCUUGUAAAUCAUAUGGCUA____UUUGC ................(((.........((((((.(((....))).....(((.(((((....))))).)))........)))))).........)))............. (-17.45 = -18.37 + 0.92)
| Location | 4,561,263 – 4,561,374 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -18.07 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
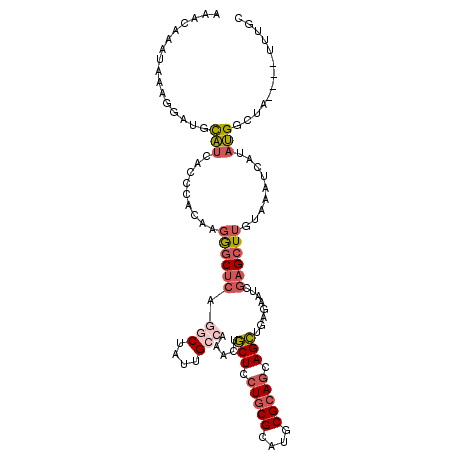
>3R_DroMel_CAF1 4561263 111 - 27905053 GCAAAUAAAUAACCAUAGGAUUUACAAGCUCGAUUGUCAGCUGCUGCGCACGCGCAGUAGCAGUUUGGCAAUAGUUUGAGCCCUUAUGGGUGAUGCAUCCUUUAUUUGCUU ((((((((........(((((...((((((..((((((((((((((((....)))))))))....))))))))))))).((((....)))).....))))))))))))).. ( -44.20) >DroSec_CAF1 18138 107 - 1 GCAAA----UAACCAUAGGAUUUACAAGCUCGAUUCUCAGCUGCUGCGCAUGCGCAGGAGCAGUUUGGCAAUAGCCUGAGCCCUUGUGGGAGAUGCAUCCUUUAUUUGUUU (((((----(((....(((((......((((....(((((((.(((((....))))).))).....(((....)))))))(((....))).)).))))))))))))))).. ( -36.30) >DroSim_CAF1 18838 107 - 1 GCAUA----UAACCAUUGGAUUUACAAGCUCGAUUCUCAGCUGCUGCGCAUGCGCAGGAGCAGUUUGGCAAUAGCCUGAGCCCUUGUGGGAGAUGCAUCCUUUAUUUGUUU ((((.----...((((.((........(((((.......(((.(((((....))))).))).....(((....))))))))))..))))...))))............... ( -34.60) >DroEre_CAF1 18579 107 - 1 GCAAA----UAGCCAUAUGAUUUACAAGCUCGAUUCUCAGCUGCUGCGCAUGCGCAGGAGCAGUUUGGCAAUAGCCUGAGCCCUAGUGGGUGAUACAUCCUUUAUUUGUUU (((((----(((....(((..((((..(((((.......(((.(((((....))))).))).....(((....))))))))((....))))))..)))...)))))))).. ( -34.10) >DroYak_CAF1 18373 107 - 1 GCAAA----UAGCCAUAUGAUUUACAAGCUCGAUUCUCAGCUGCUGCGCAUGCGCAGUAGCAAUUUGGCAAUAGCCUGAGCUCUUGUGAUUGAUACAUCCUUUAUUUGUUU (((((----(((...(((.(.(((((((((((.......(((((((((....))))))))).....(((....)))))))))..))))).).)))......)))))))).. ( -36.80) >DroAna_CAF1 15321 102 - 1 ---UA----UAGCCU-AU-UUUUGCUAUCAAACAUCUCAACUGUUUCGCAUUCGCAUGAGUAGGUUGGCAAUAGAUUGAGCCUGCCGUGGCGAAGAAUCAACUACUUGUUU ---..----((((((-((-((.(((.((((((((.......))))).).))..))).))))))))))(((((((.((((..((((....))..))..))))))).)))).. ( -23.40) >consensus GCAAA____UAACCAUAGGAUUUACAAGCUCGAUUCUCAGCUGCUGCGCAUGCGCAGGAGCAGUUUGGCAAUAGCCUGAGCCCUUGUGGGUGAUGCAUCCUUUAUUUGUUU ...................................(((((((.(((((....))))).))).....(((....)))))))(((....)))..................... (-18.07 = -18.18 + 0.12)

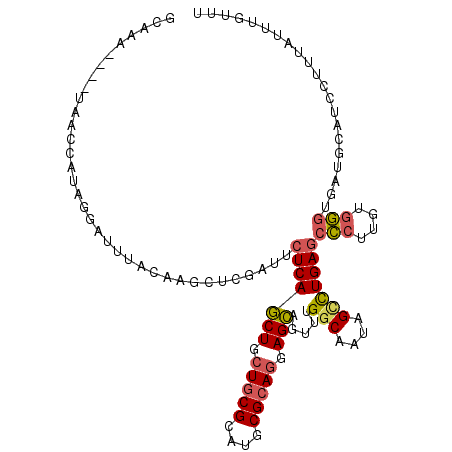
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:18 2006