
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,506,554 – 4,506,767 |
| Length | 213 |
| Max. P | 0.981893 |

| Location | 4,506,554 – 4,506,647 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.88 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4506554 93 - 27905053 GAUCGAUGGCCAUCGCCCAAGUGGAUGUUGUUCUUUCUUUGGCCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAACUAAGAAAAUCUA ....((((((....)))..(((.((.((.((.((((((((.((((((.....)))).)).)))))))).)).))))...)))......))).. ( -22.10) >DroSec_CAF1 6533 85 - 1 GAUCGAUGGCCAUCGCCCAAGUGGAUU-------UU-UUUUACCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAAUUGAGAAAAUCUU ..((((((((....)))..(((((.((-------((-((((((((((.....)))).))))))))))...)))))....)))))......... ( -22.10) >DroSim_CAF1 6534 86 - 1 GAUCGAUGGCCAUCGCCCAAGUGGAUU-------UUUUUUUGCCACUCAAAAAGUGAGUAAAAGAGAGUGCAACUCAAAAUUGAGAAAAUCUA ....((((((....)))....((.(((-------(((((((((((((.....)))).)))))))))))).)).((((....))))...))).. ( -22.30) >DroYak_CAF1 6586 84 - 1 GAUCGAUGGCCAUCGCCCAAGUGGAUU-------UCCUUUGCCCACUCAAAAAGUGAGUAAAAGAGAGUUAUACUCAAAAC--AAAAAAUCUA ....((((((....)))..((((..((-------((.(((((.((((.....)))).))))).))))....))))......--.....))).. ( -15.90) >consensus GAUCGAUGGCCAUCGCCCAAGUGGAUU_______UUCUUUGGCCACUCAAAAAGUGAGUAAAAGAGAGUGCUACUCAAAACUGAGAAAAUCUA ((((.(((((....)))...)).))))........((((((((((((.....)))).))))))))(((.....)))................. (-13.62 = -14.88 + 1.25)


| Location | 4,506,612 – 4,506,706 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -14.06 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4506612 94 + 27905053 AAGAACAACAUCCACUUGGGCGAUGGCC-----AUCGAUCCUUGUCCAC-GC-----AGGACAUUUCCGGAUUGCAUUCAAUUGUCAUCCUGCAACAAUUGCGCU ................(((((((.((..-----......)))))))))(-((-----((((....)))((((.(((......))).)))).........)))).. ( -22.60) >DroVir_CAF1 8587 85 + 1 C-------UACCCACUUGGGCGAU-----------CUAUCU-UGUCCGC-GCAGCACAGGACAUUUCCGGAUUGCAUUCAAUUGUCAUGCUGCAACAAUUGCGCU .-------.........((((((.-----------.....)-)))))..-((((((...((((......((......))...)))).))))))............ ( -21.40) >DroPse_CAF1 6636 92 + 1 A-------AAACCGCUUGGGCGAUGGCCAUCCCAUCGAUCCUUGUCCAC-GC-----AGGACACUUCCGGAUUGCAUUCAAUUGUAGUUCUGCAACAAUUGCGCU .-------.....((..(((((((((.....)))))).))).(((((..-..-----.))))).....((((((((......)))))))).(((.....))))). ( -27.70) >DroGri_CAF1 6383 86 + 1 A-------UUUCCACUUGGGCGAU-----------CCAUCU-UGUCCAUAGCGGUACAGGACAUUUCCGGAUUGCAUUCAAUUGUCAUGCUGUAACAAUUGCGCA .-------...((....))(((((-----------((....-(((((.....)).)))(((....))))))))))...(((((((.........))))))).... ( -21.30) >DroMoj_CAF1 5822 86 + 1 A-------UUUCCACUUGGGCGAU-----------CUAUCUAUGUCCAU-GCAGCAUAGGACAUUUCCGGAUUCCAUUCAAUUGUCAUAUUGCAACAAUUGUGCU .-------..........((.(((-----------((....((((((((-(...))).))))))....)))))))...(((((((.........))))))).... ( -17.30) >DroAna_CAF1 4877 87 + 1 A-------AUGUCACUUGGAUGAUGGCC-----AUCGAUCCUUGUCCAC-GC-----AGGACAUUUCCGGAUUGCAUUCAAUUGUGAUUCUGCAACAAUUGCGCU .-------..((((((((((((......-----..((((((.(((((..-..-----.))))).....)))))))))))))..)))))...(((.....)))... ( -22.20) >consensus A_______UAUCCACUUGGGCGAU___________CGAUCCUUGUCCAC_GC_____AGGACAUUUCCGGAUUGCAUUCAAUUGUCAUGCUGCAACAAUUGCGCU ...........((....))(((...............((((.(((((...........))))).....))))......(((((((.........)))))))))). (-14.06 = -13.57 + -0.50)



| Location | 4,506,647 – 4,506,744 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.75 |
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.05 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4506647 97 + 27905053 CUUGUCCAC-GC-----AGGACAUUUCCGGAUUGCAUUCAAUUGUCAUCCUGCAACAAUUGCGCUUUUCUCUGGAUUCUCCAUUUGUUUGUCU--UCUUCCGAUU ..(((((..-..-----.)))))....((((..(((..(((.((..((((((((.....)))).........))))....)).)))..)))..--...))))... ( -19.20) >DroVir_CAF1 8609 82 + 1 U-UGUCCGC-GCAGCACAGGACAUUUCCGGAUUGCAUUCAAUUGUCAUGCUGCAACAAUUGCGCUUUUUAUU-------CAUGCUGUUUGU-------------- .-....((.-((((((..(((....)))((((.((...(((((((.........))))))).)).....)))-------).)))))).)).-------------- ( -17.10) >DroGri_CAF1 6405 91 + 1 U-UGUCCAUAGCGGUACAGGACAUUUCCGGAUUGCAUUCAAUUGUCAUGCUGUAACAAUUGCGCAUUUUUCU-------A---AUGCUUGUUU---CUUCCGAUU .-(((((...........)))))....((((..(((..(((((((.........))))))).(((((.....-------)---)))).)))..---..))))... ( -19.80) >DroSim_CAF1 6620 97 + 1 CUUGUCCAC-GC-----AGGACAUUUCCGGAUUGCAUUCAAUUGUCAUCCUGCAACAAUUGCGCUUUUCUUUUGAUUCUCCAUUUGUUUGUCU--UCUUCCGAUU ..(((((..-..-----.)))))....((((..(((..(((.((.......(((.....))).....((....)).....)).)))..)))..--...))))... ( -17.10) >DroMoj_CAF1 5844 97 + 1 UAUGUCCAU-GCAGCAUAGGACAUUUCCGGAUUCCAUUCAAUUGUCAUAUUGCAACAAUUGUGCUUUUUAUU-------CCAAAUGUUUGUUUUUUUUUCGGAUU .((((((((-(...))).)))))).((((((...((..((.(((..(((..(((.(....))))....))).-------.))).))..)).......)))))).. ( -18.20) >DroAna_CAF1 4905 96 + 1 CUUGUCCAC-GC-----AGGACAUUUCCGGAUUGCAUUCAAUUGUGAUUCUGCAACAAUUGCGCUUUUCUUCACAUUUUCCAUUUGUCUGUCU---CUUCCUAUU ..(((((..-..-----.)))))....(((((..........(((((...((((.....)))).......)))))..........)))))...---......... ( -16.35) >consensus CUUGUCCAC_GC_____AGGACAUUUCCGGAUUGCAUUCAAUUGUCAUCCUGCAACAAUUGCGCUUUUCUUU_______CCAUUUGUUUGUCU___CUUCCGAUU ..(((((...........))))).....(((..((...(((((((.........))))))).))..))).................................... (-11.13 = -11.05 + -0.08)



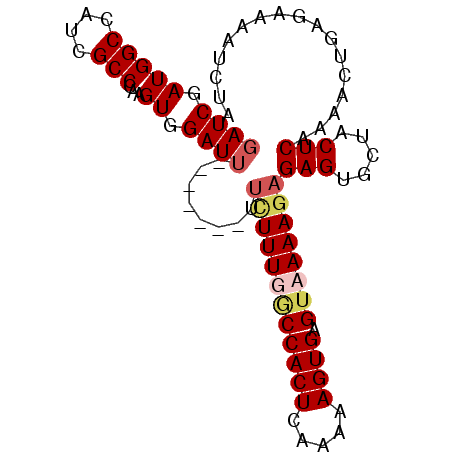
| Location | 4,506,647 – 4,506,744 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.75 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -12.95 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4506647 97 - 27905053 AAUCGGAAGA--AGACAAACAAAUGGAGAAUCCAGAGAAAAGCGCAAUUGUUGCAGGAUGACAAUUGAAUGCAAUCCGGAAAUGUCCU-----GC-GUGGACAAG ..(((((...--...........(((.....))).......((.((((((((.......))))))))...))..)))))...(((((.-----..-..))))).. ( -24.80) >DroVir_CAF1 8609 82 - 1 --------------ACAAACAGCAUG-------AAUAAAAAGCGCAAUUGUUGCAGCAUGACAAUUGAAUGCAAUCCGGAAAUGUCCUGUGCUGC-GCGGACA-A --------------.....((((((.-------........((.((((((((.......))))))))...)).....(((....))).)))))).-.......-. ( -18.80) >DroGri_CAF1 6405 91 - 1 AAUCGGAAG---AAACAAGCAU---U-------AGAAAAAUGCGCAAUUGUUACAGCAUGACAAUUGAAUGCAAUCCGGAAAUGUCCUGUACCGCUAUGGACA-A ..(((((..---......((((---(-------.....))))).(((((((((.....))))))))).......)))))...(((((...........)))))-. ( -23.90) >DroSim_CAF1 6620 97 - 1 AAUCGGAAGA--AGACAAACAAAUGGAGAAUCAAAAGAAAAGCGCAAUUGUUGCAGGAUGACAAUUGAAUGCAAUCCGGAAAUGUCCU-----GC-GUGGACAAG ..((....))--...........((((...((....))...((.((((((((.......))))))))...))..))))....(((((.-----..-..))))).. ( -21.30) >DroMoj_CAF1 5844 97 - 1 AAUCCGAAAAAAAAACAAACAUUUGG-------AAUAAAAAGCACAAUUGUUGCAAUAUGACAAUUGAAUGGAAUCCGGAAAUGUCCUAUGCUGC-AUGGACAUA ..((((.........(((....))).-------...........((((((((.......)))))))).........)))).((((((.(((...)-)))))))). ( -19.40) >DroAna_CAF1 4905 96 - 1 AAUAGGAAG---AGACAGACAAAUGGAAAAUGUGAAGAAAAGCGCAAUUGUUGCAGAAUCACAAUUGAAUGCAAUCCGGAAAUGUCCU-----GC-GUGGACAAG .........---...........((((...((((........))))....(((((...(((....))).)))))))))....(((((.-----..-..))))).. ( -17.90) >consensus AAUCGGAAG___AGACAAACAAAUGG_______AAAAAAAAGCGCAAUUGUUGCAGCAUGACAAUUGAAUGCAAUCCGGAAAUGUCCU_____GC_GUGGACAAA .........................................(((((((((((.......))))))))..)))..........(((((...........))))).. (-12.95 = -13.15 + 0.20)



| Location | 4,506,675 – 4,506,767 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Mean single sequence MFE | -15.66 |
| Consensus MFE | -6.19 |
| Energy contribution | -6.25 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4506675 92 - 27905053 AGAAG-------UAAGAAAAUGUGACAGGCAAUCGGAAGA--AGACAAACAAAUGGAGAAUCCAGAGAAAAGCGCAAUUGUUGCAGGAUGACAAUUGAAUG ....(-------(.......((..(((.((..((....))--...........(((.....))).........))...)))..)).....))......... ( -15.10) >DroGri_CAF1 6438 88 - 1 AACCAUUUGUAUGAAGAAAAGGUGACAAACAAUCGGAAG---AAACAAGCAU---U-------AGAAAAAUGCGCAAUUGUUACAGCAUGACAAUUGAAUG ......((((..........(....)......((....)---).))))((((---(-------.....))))).(((((((((.....))))))))).... ( -17.60) >DroMoj_CAF1 5877 93 - 1 AACCAUUGCCC-UAAGAAAAGGUGACAAGCAAUCCGAAAAAAAAACAAACAUUUGG-------AAUAAAAAGCACAAUUGUUGCAAUAUGACAAUUGAAUG ......(((..-........(....)......((((((.............)))))-------).......)))((((((((.......)))))))).... ( -14.22) >DroAna_CAF1 4933 89 - 1 ACAAG-------AAAC--AAUGUGACAGGCAAUAGGAAG---AGACAGACAAAUGGAAAAUGUGAAGAAAAGCGCAAUUGUUGCAGAAUCACAAUUGAAUG .....-------...(--(((((((...(((((((....---...((......)).....((((........)))).)))))))....)))).)))).... ( -15.70) >consensus AAAAA_______UAAGAAAAGGUGACAAGCAAUCGGAAG___AAACAAACAAAUGG_______AAAAAAAAGCGCAAUUGUUGCAGAAUGACAAUUGAAUG ..........................................................................(((((((((.....))))))))).... ( -6.19 = -6.25 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:00 2006