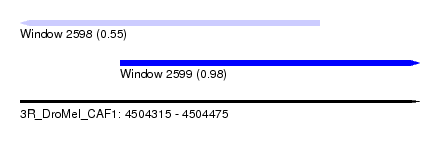
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,504,315 – 4,504,475 |
| Length | 160 |
| Max. P | 0.984435 |

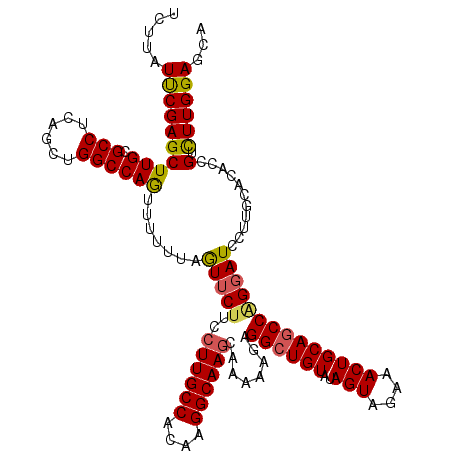
| Location | 4,504,315 – 4,504,435 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4504315 120 - 27905053 UCUUAUUCGAGCUUGAGCCUCAGGUGGCCAGUUUUUUAAUUCUUCCUUGCCACAAGGCAAGCAAAAAGAGGCUGUAUAGUAGAAACUGCAGCCAGGAUCCUUGCACACCGUCUUGGAGCA ..........((((.((.....((((((.((..((((...((((.((((((....))))))....))))((((((..(((....)))))))))))))..)).)).))))...)).)))). ( -37.10) >DroSec_CAF1 4295 120 - 1 UCUUAUUCGAGCUUGGGCCUCAGCUGGCCAAUUUUUUAGUUCUUCCUUGCCACAAGGCAAACAAAAAGAGGCUGUAUAGUAGAAACUGCAGCCAGGAUCCUUGCACACCGUCUUGGAGCA ........(((((..((((......))))........))))).....(((..((((((...........((((((..(((....))))))))).((...........))))))))..))) ( -34.10) >DroSim_CAF1 4290 120 - 1 UCUUAUUCGAGCUUGGGCCUCAGCUGGCCAAUUUUUUAGUUCUUCCUUGCCACAAGGCAAGCAAAAAGAGGCUGUGUAGUAGAAACUGCAGCCAGGAUCCUUGCACACCGUCUUGGAGCA ...............((((......)))).........(((((..((((((....))))))......(((((.((((.((((...)))).((.((....)).)))))).)))))))))). ( -36.70) >DroEre_CAF1 4298 120 - 1 UCGCAUUCGAGCUUGCGCCUCAACUGGCCAGUUCUUUAGUUCCACCUUGCCACAAGGCAAGCAAAAUGAGGAUGUAUAGUAAAAACUGCAGCCAGUAUCCUUGCAUACGGAUUUGGAGCA ........(((((.(.(((......)))))))))....(((((..((((((....))))))......(((..(((((.((((..((((....))))....)))))))))..)))))))). ( -34.20) >DroYak_CAF1 4309 120 - 1 UCUUAUCCGAGCUUGCGCCUCAACUGGCCAGUUCUUUAGUUCUUCCUUGCCACAAGGCAAGCAAAAAGAGGCUGUGUAGUAAAAACUGCAGCCGGGAUCCUUGCACACUGACUUGGAGCA .....(((((((.((((......((((((((((((((.....((.((((((....)))))).))))))).))))((((((....)))))))))))......))))....).))))))... ( -37.10) >consensus UCUUAUUCGAGCUUGCGCCUCAGCUGGCCAGUUUUUUAGUUCUUCCUUGCCACAAGGCAAGCAAAAAGAGGCUGUAUAGUAGAAACUGCAGCCAGGAUCCUUGCACACCGUCUUGGAGCA .....((((((((((.(((......)))))).......(((((..((((((....))))))........((((((..(((....))))))))))))))...........).))))))... (-28.26 = -28.22 + -0.04)


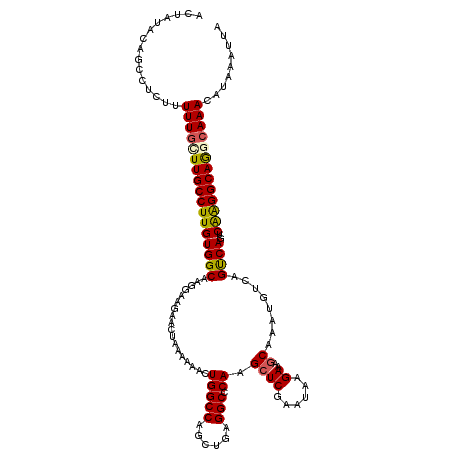
| Location | 4,504,355 – 4,504,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.28 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4504355 120 + 27905053 ACUAUACAGCCUCUUUUUGCUUGCCUUGUGGCAAGGAAGAAUUAAAAAACUGGCCACCUGAGGCUCAAGCUCGAAUAAGAAAACAAAUGUCUGUCAGUCAAGGCAGGCAAACAUAAAUUA .......(((((((((((.((((((....)))))).)))))..........((...)).))))))......................((((((((......))))))))........... ( -32.70) >DroSec_CAF1 4335 120 + 1 ACUAUACAGCCUCUUUUUGUUUGCCUUGUGGCAAGGAAGAACUAAAAAAUUGGCCAGCUGAGGCCCAAGCUCGAAUAAGAAAGCAAAUGUCAGCCAGUCAGGGCAGGCAAACAUAAAUUA ...............(((((((((((((((((...................((((......))))...((((......))..))........))))..)))))))))))))......... ( -32.20) >DroSim_CAF1 4330 120 + 1 ACUACACAGCCUCUUUUUGCUUGCCUUGUGGCAAGGAAGAACUAAAAAAUUGGCCAGCUGAGGCCCAAGCUCGAAUAAGAAAGCAAAUGUCAGUCAGUCAGGGCAGGCAAACAUGAAUUA ...........((..(((((((((((((((((...................((((......))))...((((......))..))........))))..)))))))))))))...)).... ( -32.30) >DroEre_CAF1 4338 120 + 1 ACUAUACAUCCUCAUUUUGCUUGCCUUGUGGCAAGGUGGAACUAAAGAACUGGCCAGUUGAGGCGCAAGCUCGAAUGCGAAAGCAAUUGUCAGUCAGUCGAGGCAGCCAAACAUAAAUUA .........((((.(((..((((((....))))).)..))).....((.(((((...(((((.(....)))))).(((....))).......)))))))))))................. ( -36.40) >DroYak_CAF1 4349 120 + 1 ACUACACAGCCUCUUUUUGCUUGCCUUGUGGCAAGGAAGAACUAAAGAACUGGCCAGUUGAGGCGCAAGCUCGGAUAAGAAAGCAGUUGUCAGCCAGUCAAGGCAAAAAAAAUUAAAAAA ........((((.(((((.((((((....)))))).))))).....((.(((((....((((.(....)))))(((((........))))).))))))).))))................ ( -36.60) >consensus ACUAUACAGCCUCUUUUUGCUUGCCUUGUGGCAAGGAAGAACUAAAAAACUGGCCAGCUGAGGCCCAAGCUCGAAUAAGAAAGCAAAUGUCAGUCAGUCAAGGCAGGCAAACAUAAAUUA ...............(((((((((((((((((..................(((((......))).)).((((......))..))........))))..)))))))))))))......... (-27.36 = -27.28 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:54 2006