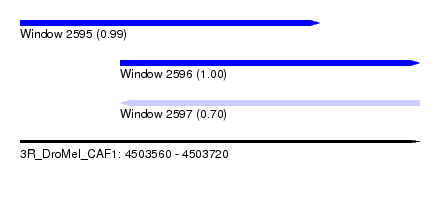
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,503,560 – 4,503,720 |
| Length | 160 |
| Max. P | 0.999946 |

| Location | 4,503,560 – 4,503,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -42.28 |
| Energy contribution | -41.64 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503560 120 + 27905053 CUUCCACAAGGGCUGCUAUGUGGGCCAGGAGCUAACUGCCCGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCAAUUGAUGUGGGUUCCAGCCA ..((((((.(((((.....((((((.((.......))))))))(((.(((....(((((((((((....).))).)))))))....))).))))))))(....).))))))......... ( -48.80) >DroSec_CAF1 3542 120 + 1 CUUCCACAAGGGCUGCUAUGUGGGCCAGGAGCUAACAGCCCGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCA ..........(((((..((..(((((.((.((.....))))..(((.(((....(((((((((((....).))).)))))))....))).))))))))((((....))))))..))))). ( -48.20) >DroSim_CAF1 3539 120 + 1 CUUCCACAAGGGCUGCUAUGUGGGCCAGGAGCUAACAGCCCGCGUCCAUCAUUCGGGCGUGAUACGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCA ..........(((((..((..(((((.((.((.....))))..(((.(((....(((((((((.((....)))).)))))))....))).))))))))((((....))))))..))))). ( -44.50) >DroEre_CAF1 3539 120 + 1 CUUCCAAAAGGGCUGCUAUGUGGGCCAGGAGUUGACUGCCCGCGUUCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCUCCUAUUGAUUUGGGUUCCAGCCA ..........((((.....((((((.((.......))))))))(((.(((....(((((((((((....).))).)))))))....))).)))((..((((......))))..)))))). ( -46.10) >DroYak_CAF1 3552 120 + 1 CUUCCAAAAGGGCUGCUAUGUCGGCCAGGAGUUGACCGCCCGCGUCCAUCAUUCGGGCGUGAUCCGAAAGAGAUACAUGCCCAUCCGGUUGACAGCUCCUAUUGAUUUGGGUUCCAACCA ..((((((..(((((......)))))((((((((.(.(((...(.....)....(((((((((((....).))).)))))))....))).).)))))))).....))))))......... ( -44.00) >consensus CUUCCACAAGGGCUGCUAUGUGGGCCAGGAGCUAACAGCCCGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCA ..((((((.(((((.....((((((............))))))(((.(((....(((((((((((....).))).)))))))....))).))))))))(....).))))))......... (-42.28 = -41.64 + -0.64)


| Location | 4,503,600 – 4,503,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -55.14 |
| Consensus MFE | -50.44 |
| Energy contribution | -51.28 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503600 120 + 27905053 CGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCAAUUGAUGUGGGUUCCAGCCAGGACGUAACUUCUUUGGCUGGAGCGAAACUUGGACGCGUC ((((((((...((((((((((((((....).))).)))))))..(((.....)))..(((.......)))(((((((((((((.......)).))))))))))))))...)))))))).. ( -57.60) >DroSec_CAF1 3582 120 + 1 CGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCAGGACGUAACUUCUUUGGCUGGAGCGAAACUUGGACGCGUC ((((((((...((((((((((((((....).))).)))))))..(((.....)))...((((....))))(((((((((((((.......)).))))))))))))))...)))))))).. ( -57.50) >DroSim_CAF1 3579 120 + 1 CGCGUCCAUCAUUCGGGCGUGAUACGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCAGGACGUAACUUCUUUGGCUGGAGCGAAACUUGGACGCGUC ((((((((...((((((((((((.((....)))).)))))))..(((.....)))...((((....))))(((((((((((((.......)).))))))))))))))...)))))))).. ( -53.80) >DroEre_CAF1 3579 120 + 1 CGCGUUCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCUCCUAUUGAUUUGGGUUCCAGCCAGGAUGUAACUUCUGUGGCUGGAGCGAAACUUGGACGCGUC ((((((((...((((((((((((((....).))).)))))))..(((.....)))...............(((((((((((((.......))).)))))))))))))...)))))))).. ( -54.60) >DroYak_CAF1 3592 120 + 1 CGCGUCCAUCAUUCGGGCGUGAUCCGAAAGAGAUACAUGCCCAUCCGGUUGACAGCUCCUAUUGAUUUGGGUUCCAACCAGGACGUAACUUCUGUUGCUGGAGCGAAACUAGGACGCGUC (((((((.......(((((((((((....).))).)))))))....((((....(((((......(((((.......)))))..(((((....))))).)))))..)))).))))))).. ( -52.20) >consensus CGCGUCCAUCAUUCGGGCGUGAUCCGCAAGCGAUACAUGCCCAUCCGGUUGACGGCCCCUAUUGAUGUGGGUUCCAGCCAGGACGUAACUUCUUUGGCUGGAGCGAAACUUGGACGCGUC ((((((((...((((((((((((((....).))).)))))))..(((.....)))...............(((((((((((((.......))).)))))))))))))...)))))))).. (-50.44 = -51.28 + 0.84)



| Location | 4,503,600 – 4,503,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -37.76 |
| Energy contribution | -38.64 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503600 120 - 27905053 GACGCGUCCAAGUUUCGCUCCAGCCAAAGAAGUUACGUCCUGGCUGGAACCCACAUCAAUUGGGGCCGUCAACCGGAUGGGCAUGUAUCGCUUGCGGAUCACGCCCGAAUGAUGGACGCG ..((((((((...(((..((((((((..((.......)).)))))))).((((.......)))).(((.....)))..((((.((..(((....)))..)).)))))))...)))))))) ( -47.20) >DroSec_CAF1 3582 120 - 1 GACGCGUCCAAGUUUCGCUCCAGCCAAAGAAGUUACGUCCUGGCUGGAACCCACAUCAAUAGGGGCCGUCAACCGGAUGGGCAUGUAUCGCUUGCGGAUCACGCCCGAAUGAUGGACGCG ..((((((((...(((..((((((((..((.......)).)))))))).(((.........))).(((.....)))..((((.((..(((....)))..)).)))))))...)))))))) ( -45.90) >DroSim_CAF1 3579 120 - 1 GACGCGUCCAAGUUUCGCUCCAGCCAAAGAAGUUACGUCCUGGCUGGAACCCACAUCAAUAGGGGCCGUCAACCGGAUGGGCAUGUAUCGCUUGCGUAUCACGCCCGAAUGAUGGACGCG ..((((((((...(((..((((((((..((.......)).)))))))).(((.........))).(((.....)))..((((.((...((....))...)).)))))))...)))))))) ( -45.90) >DroEre_CAF1 3579 120 - 1 GACGCGUCCAAGUUUCGCUCCAGCCACAGAAGUUACAUCCUGGCUGGAACCCAAAUCAAUAGGAGCCGUCAACCGGAUGGGCAUGUAUCGCUUGCGGAUCACGCCCGAAUGAUGAACGCG ..(((((.((...(((..((((((((..((.......)).)))))))).................(((.....)))..((((.((..(((....)))..)).)))))))...)).))))) ( -36.10) >DroYak_CAF1 3592 120 - 1 GACGCGUCCUAGUUUCGCUCCAGCAACAGAAGUUACGUCCUGGUUGGAACCCAAAUCAAUAGGAGCUGUCAACCGGAUGGGCAUGUAUCUCUUUCGGAUCACGCCCGAAUGAUGGACGCG ..(((((((....(((((....))............((((.((((((..(((.........)).)...))))))))))((((.((.((((.....)))))).)))))))....))))))) ( -40.40) >consensus GACGCGUCCAAGUUUCGCUCCAGCCAAAGAAGUUACGUCCUGGCUGGAACCCACAUCAAUAGGGGCCGUCAACCGGAUGGGCAUGUAUCGCUUGCGGAUCACGCCCGAAUGAUGGACGCG ..((((((((...(((..((((((((..((.......)).)))))))).(((.........))).(((.....)))..((((.((...((....))...)).)))))))...)))))))) (-37.76 = -38.64 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:52 2006