
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,503,047 – 4,503,207 |
| Length | 160 |
| Max. P | 0.999931 |

| Location | 4,503,047 – 4,503,167 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -35.76 |
| Energy contribution | -37.04 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503047 120 + 27905053 UCAGUCGCCGGGGGGUCCUGCCUCAAUGUACGCCCAUUUUCUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGA ....((((..(((.((.(.........).)).))).....((((((((((.(((..((.(((.((((((.......)))))).))))).....)))(....)...)))))))))).)))) ( -40.80) >DroSec_CAF1 3029 120 + 1 UCAGUCGCCGGGGGGUCCUGCCUCAAUGUACGCCCACUUUCUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGA ..(((.(((..((((.....)))).......((((.((((......)))).)))).))).)))((((((.......))))))(((((((((((...(((.....))))))))).))))). ( -43.30) >DroSim_CAF1 3026 120 + 1 UCAGUCGCCGGGGGGUCCUGCCUCAAUGUACGCCCACUUUCUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGA ..(((.(((..((((.....)))).......((((.((((......)))).)))).))).)))((((((.......))))))(((((((((((...(((.....))))))))).))))). ( -43.30) >DroEre_CAF1 3026 120 + 1 UCGGUCGCCGGGAGGUCCUGCCUCAAUGUACGCCCACUUCCUCAACAAGGCGGGCAGGCUUCUGUAUGACACAAUCAUGUACCGCACCAACAACCCAGAAACCAUCCUAGUUGAGUGCGA .(((..(((..((((.....)))).......((((.(((........))).)))).)))..)))(((((.....)))))...(((((((((..................)))).))))). ( -35.57) >DroYak_CAF1 3039 120 + 1 UCGUUCACCGGGGGGUCCUGCCUCAAUGUACGCCCACUUCCUCAACAAGGCGGGCAGGCUGCUGUACGACACAAUCUUGUACCGCACCAACAAUCCGGAAACCAUCCUAAUUGAGUGCGA ((((.(.((((((((.....))))..(((..((((.(((........))).)))).((.(((.((((((.......)))))).))))).)))..))))....((.......)).).)))) ( -36.20) >consensus UCAGUCGCCGGGGGGUCCUGCCUCAAUGUACGCCCACUUUCUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGA ..(((.(((..((((.....)))).......((((.((((......)))).)))).))).)))((((((.......))))))(((((((((((...(((.....))))))))).))))). (-35.76 = -37.04 + 1.28)

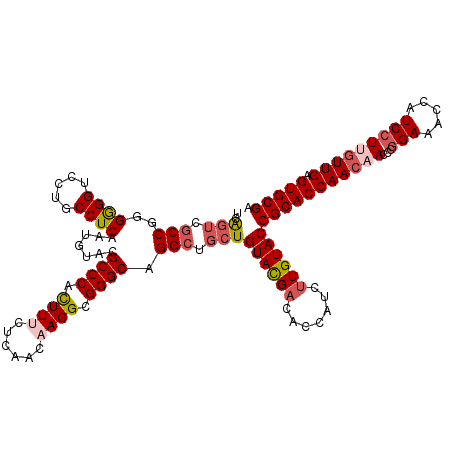

| Location | 4,503,047 – 4,503,167 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -47.54 |
| Consensus MFE | -45.38 |
| Energy contribution | -45.74 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503047 120 - 27905053 UCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAGAAAAUGGGCGUACAUUGAGGCAGGACCCCCCGGCGACUGA ((((.((((((((((.(((....((((((((((.((((..((......))..)))))))))))))).))))))))))))).....(((.((...(((...))).)))))...)))).... ( -51.20) >DroSec_CAF1 3029 120 - 1 UCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAGAAAGUGGGCGUACAUUGAGGCAGGACCCCCCGGCGACUGA ((((.((((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))))...(.(((.((...(((...))).)).)))).)))).... ( -51.80) >DroSim_CAF1 3026 120 - 1 UCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAGAAAGUGGGCGUACAUUGAGGCAGGACCCCCCGGCGACUGA ((((.((((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))))...(.(((.((...(((...))).)).)))).)))).... ( -51.80) >DroEre_CAF1 3026 120 - 1 UCGCACUCAACUAGGAUGGUUUCUGGGUUGUUGGUGCGGUACAUGAUUGUGUCAUACAGAAGCCUGCCCGCCUUGUUGAGGAAGUGGGCGUACAUUGAGGCAGGACCUCCCGGCGACCGA .(((.((((((.(((..((((((((......(((..((((.....))))..)))..))))))))......))).))))))...)))(((((.....((((.....))))...))).)).. ( -41.20) >DroYak_CAF1 3039 120 - 1 UCGCACUCAAUUAGGAUGGUUUCCGGAUUGUUGGUGCGGUACAAGAUUGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAGGAAGUGGGCGUACAUUGAGGCAGGACCCCCCGGUGAACGA ((((.((((((.(((..(((....((.((((((.((((..(((....)))..)))))))))))).)))..))).))))))...)))).(((.(((((.((........))))))).))). ( -41.70) >consensus UCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAGAAAGUGGGCGUACAUUGAGGCAGGACCCCCCGGCGACUGA ((((.((((((((((.(((....((((((((((.((((..((......))..)))))))))))))).))))))))))))).....(((.((...(((...))).)).)))..)))).... (-45.38 = -45.74 + 0.36)



| Location | 4,503,087 – 4,503,207 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -44.14 |
| Consensus MFE | -41.38 |
| Energy contribution | -42.34 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503087 120 + 27905053 CUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUCCGACGCCAUCUUCGCACC .(((((((((.(((..((.(((.((((((.......)))))).))))).....)))(....)...)))))))))((((((..((..(((....((((....)))).)))..)))))))). ( -47.20) >DroSec_CAF1 3069 120 + 1 CUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUCCGACGCCACCUGCGCACC .(((((((((.(((..((.(((.((((((.......)))))).))))).....)))(....)...)))))))))(((((..((((.(((....((((....)))).))).))))))))). ( -49.60) >DroSim_CAF1 3066 120 + 1 CUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUCCGACGCCACCUGCGCACC .(((((((((.(((..((.(((.((((((.......)))))).))))).....)))(....)...)))))))))(((((..((((.(((....((((....)))).))).))))))))). ( -49.60) >DroEre_CAF1 3066 120 + 1 CUCAACAAGGCGGGCAGGCUUCUGUAUGACACAAUCAUGUACCGCACCAACAACCCAGAAACCAUCCUAGUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUUCGACGCCACCUGCGCACU .(((((.((((((((((....))))((((.....))))...))))............((.....)))).)))))(((((..((((.(((....((((....)))).))).))))))))). ( -34.30) >DroYak_CAF1 3079 120 + 1 CUCAACAAGGCGGGCAGGCUGCUGUACGACACAAUCUUGUACCGCACCAACAAUCCGGAAACCAUCCUAAUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUCCGACGCCACCUCCGCACU .........(((((..((((((.((((((.......)))))).)))..........((((.((.(((..((((....))))..))))).))))((((....)))).)))...)))))... ( -40.00) >consensus CUCAACAAGGCGGGCAGGCUGCUGUACGACACCAUCUUGUACCGCACCAACAACCCGGAAACCAUCCUUGUUGAGUGCGAUCGGGAGGCUUCCUCGGACUUCCGACGCCACCUGCGCACC .(((((((((.(((..((.(((.((((((.......)))))).))))).....)))(....)...)))))))))(((((..((((.(((....((((....)))).))).))))))))). (-41.38 = -42.34 + 0.96)



| Location | 4,503,087 – 4,503,207 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -53.94 |
| Consensus MFE | -50.94 |
| Energy contribution | -52.02 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4503087 120 - 27905053 GGUGCGAAGAUGGCGUCGGAAGUCCGAGGAAGCCUCCCGAUCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAG .((((((.((.(((.((((....))))....)))))....))))))(((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))). ( -57.20) >DroSec_CAF1 3069 120 - 1 GGUGCGCAGGUGGCGUCGGAAGUCCGAGGAAGCCUCCCGAUCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAG .(((((..((.(((.((((....))))....))).))....)))))(((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))). ( -58.70) >DroSim_CAF1 3066 120 - 1 GGUGCGCAGGUGGCGUCGGAAGUCCGAGGAAGCCUCCCGAUCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAG .(((((..((.(((.((((....))))....))).))....)))))(((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))). ( -58.70) >DroEre_CAF1 3066 120 - 1 AGUGCGCAGGUGGCGUCGAAAGUCCGAGGAAGCCUCCCGAUCGCACUCAACUAGGAUGGUUUCUGGGUUGUUGGUGCGGUACAUGAUUGUGUCAUACAGAAGCCUGCCCGCCUUGUUGAG .(((((..((.(((.(((......)))....))).))....)))))(((((.(((..((((((((......(((..((((.....))))..)))..))))))))......))).))))). ( -42.70) >DroYak_CAF1 3079 120 - 1 AGUGCGGAGGUGGCGUCGGAAGUCCGAGGAAGCCUCCCGAUCGCACUCAAUUAGGAUGGUUUCCGGAUUGUUGGUGCGGUACAAGAUUGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAG .((((((((((..(.((((....)))).)..)))))).....))))(((((.(((..(((....((.((((((.((((..(((....)))..)))))))))))).)))..))).))))). ( -52.40) >consensus GGUGCGCAGGUGGCGUCGGAAGUCCGAGGAAGCCUCCCGAUCGCACUCAACAAGGAUGGUUUCCGGGUUGUUGGUGCGGUACAAGAUGGUGUCGUACAGCAGCCUGCCCGCCUUGUUGAG .(((((..((.(((.((((....))))....))).))....)))))(((((((((.(((....((((((((((.((((..((......))..)))))))))))))).)))))))))))). (-50.94 = -52.02 + 1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:48 2006