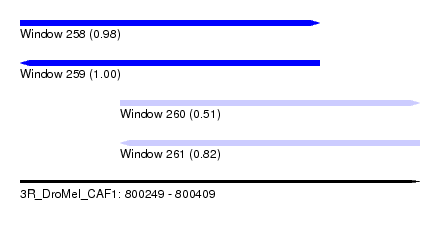
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 800,249 – 800,409 |
| Length | 160 |
| Max. P | 0.998353 |

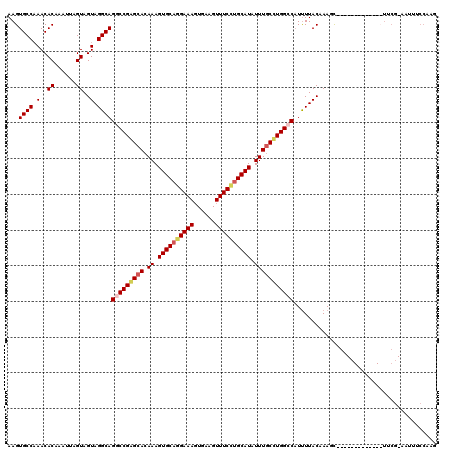
| Location | 800,249 – 800,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -25.91 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 800249 120 + 27905053 CUUGGAAAUUUCGAAGGCAUCGCGAUCGGCUUGGUAAAAUGGCCAAGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGCCUGCCUACUACUAAUUUGUGUUUGGCACUU ..............((((..(((((...(((((((......))))))).....(((((((((......)))))))))..)))))....))))(((...(((......)))...))).... ( -39.40) >DroSec_CAF1 38278 120 + 1 CUUGGAAACUUCGAAGGCAUGGCGAUCGACUUUGUAAAAUGGCCAAGCAAAUAUGCAGGAAACUUCACUUUCUUGCACUUUGUGCUCGGCCUGCCUACUACUAAUUUGUGUUUGGCACUU ...(....).....(((((..((((......)))).....((((.((((....(((((((((......))))))))).....)))).)))))))))...........(((.....))).. ( -35.40) >DroEre_CAF1 21874 106 + 1 CUUGGCAUUU-GGGA-------------GAUUUGUAAAAUGGCCAGGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGACUGCCUACUACUAAUUUGUGUUUGGCACUU ((.(((((..-((..-------------.((((((...........)))))).(((((((((......)))))))))))..))))).))..((((...(((......)))...))))... ( -29.80) >DroYak_CAF1 27734 92 + 1 ----------------------------AUUUUGUAAGAAGGCCAGGAAAAUAUGCAAGAAACUUCAGUUUCCAGCACUUUGUGCCCGGACUGCCUACUACUAAUUUGUGUUAGGCACUU ----------------------------............((((.((..((..(((..(((((....)))))..)))..))...)).)).))(((((.(((......))).))))).... ( -23.80) >consensus CUUGGAAAUU_CGAA_____________ACUUUGUAAAAUGGCCAAGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGACUGCCUACUACUAAUUUGUGUUUGGCACUU ........................................((((.((((....(((((((((......))))))))).....)))).))))((((...(((......)))...))))... (-25.91 = -26.60 + 0.69)

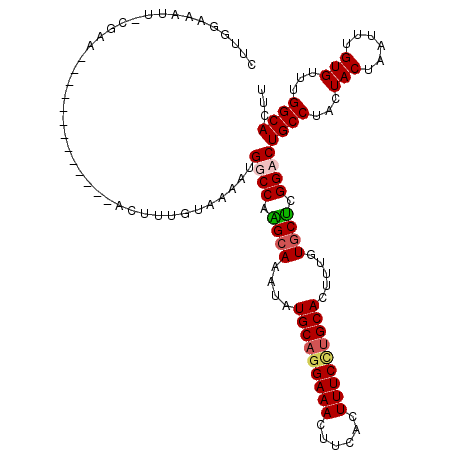
| Location | 800,249 – 800,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -28.20 |
| Energy contribution | -29.32 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 800249 120 - 27905053 AAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCUUGGCCAUUUUACCAAGCCGAUCGCGAUGCCUUCGAAAUUUCCAAG ..(((.....))).........(.((((((((((((((.((.((((((((((......)))))))))).)))))))))))..........((.....))..))))).)............ ( -41.90) >DroSec_CAF1 38278 120 - 1 AAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAAGAAAGUGAAGUUUCCUGCAUAUUUGCUUGGCCAUUUUACAAAGUCGAUCGCCAUGCCUUCGAAGUUUCCAAG ...((((.(..((......))..).))))(((((((((.((.(((((.((((......)))).))))).)))))))))))...........((((..((...))..)))).......... ( -34.70) >DroEre_CAF1 21874 106 - 1 AAGUGCCAAACACAAAUUAGUAGUAGGCAGUCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCCUGGCCAUUUUACAAAUC-------------UCCC-AAAUGCCAAG ..(((.....)))............((((.(((..((((...)))).)))..(((((((..((.(((....)))..))..))))))).....-------------....-...))))... ( -26.00) >DroYak_CAF1 27734 92 - 1 AAGUGCCUAACACAAAUUAGUAGUAGGCAGUCCGGGCACAAAGUGCUGGAAACUGAAGUUUCUUGCAUAUUUUCCUGGCCUUCUUACAAAAU---------------------------- ...((((((..((......))..))))))(.(((((...((.((((.((((((....)))))).)))).))..))))).)............---------------------------- ( -27.20) >consensus AAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCCUGGCCAUUUUACAAAGC_____________UUCG_AAUUUCCAAG ...((((.(..((......))..).))))(((((((((.((.((((((((((......)))))))))).)))))))))))........................................ (-28.20 = -29.32 + 1.12)

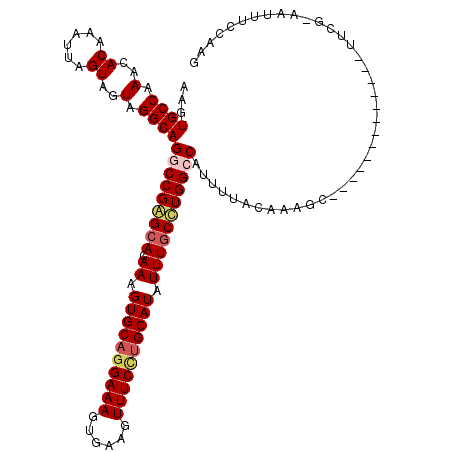
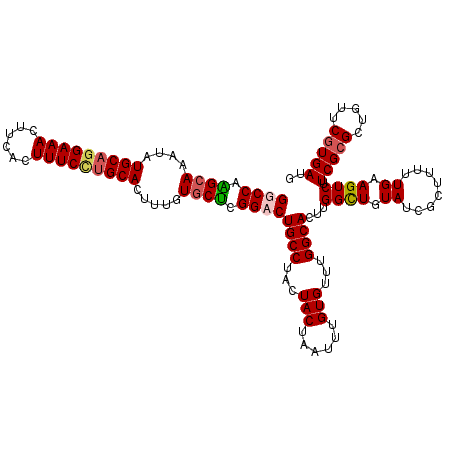
| Location | 800,289 – 800,409 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -29.25 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 800289 120 + 27905053 GGCCAAGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGCCUGCCUACUACUAAUUUGUGUUUGGCACUUGGCUGUAUCGCUUUUUGAAGUCUUCGCGCUGUUCGUGAUG ((((.((((....(((((((((......))))))))).....)))).))))((((...(((......)))...))))...((((.((........)).)))).(((((.....))))).. ( -39.20) >DroSec_CAF1 38318 120 + 1 GGCCAAGCAAAUAUGCAGGAAACUUCACUUUCUUGCACUUUGUGCUCGGCCUGCCUACUACUAAUUUGUGUUUGGCACUUGGCUGUAUCGCUUUUUGAAGUCUUCGCGCUGUUCGUGAUG ((((.((((....(((((((((......))))))))).....)))).))))((((...(((......)))...))))...((((.((........)).)))).(((((.....))))).. ( -36.50) >DroEre_CAF1 21900 119 + 1 GGCCAGGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGACUGCCUACUACUAAUUUGUGUUUGGCACUUGGCUGUAUCG-UUUUUGAAGUCUUCGCGCUGUUCGUGAUG (((((((((....(((((((((......))))))))).....)))).....((((...(((......)))...))))..)))))......-............(((((.....))))).. ( -33.90) >DroYak_CAF1 27746 120 + 1 GGCCAGGAAAAUAUGCAAGAAACUUCAGUUUCCAGCACUUUGUGCCCGGACUGCCUACUACUAAUUUGUGUUAGGCACUUGGUUGUAUCUCUUUUUGAAGUCUUCGCGCUGUUCAUGAUA ...((.(((.....((..((((((((((..(((.((((...))))..))).((((((.(((......))).)))))).................))))))).)))..))..))).))... ( -28.90) >consensus GGCCAAGCAAAUAUGCAGGAAACUUCACUUUCCUGCACUUUGUGCUCGGACUGCCUACUACUAAUUUGUGUUUGGCACUUGGCUGUAUCGCUUUUUGAAGUCUUCGCGCUGUUCGUGAUG ((((.((((....(((((((((......))))))))).....)))).))))((((...(((......)))...))))...((((.((........)).)))).(((((.....))))).. (-29.25 = -30.00 + 0.75)

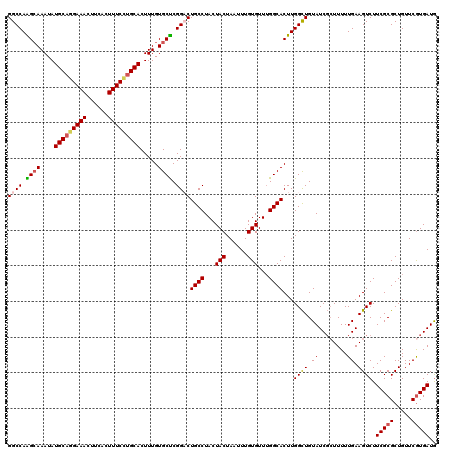
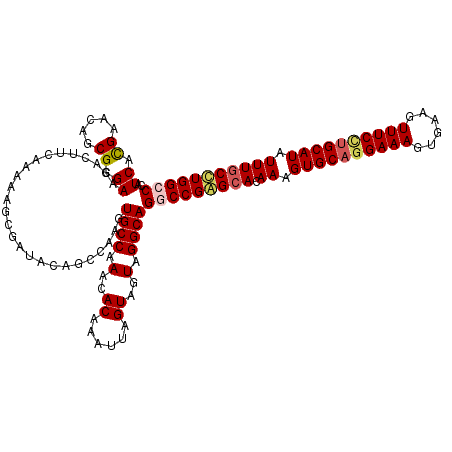
| Location | 800,289 – 800,409 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -32.59 |
| Consensus MFE | -27.69 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 800289 120 - 27905053 CAUCACGAACAGCGCGAAGACUUCAAAAAGCGAUACAGCCAAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCUUGGCC ............(((..............))).....(((.(.(((.............))).).))).(((((((((.((.((((((((((......)))))))))).))))))))))) ( -38.26) >DroSec_CAF1 38318 120 - 1 CAUCACGAACAGCGCGAAGACUUCAAAAAGCGAUACAGCCAAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAAGAAAGUGAAGUUUCCUGCAUAUUUGCUUGGCC ............(((..............))).....(((.(.(((.............))).).))).(((((((((.((.(((((.((((......)))).))))).))))))))))) ( -31.76) >DroEre_CAF1 21900 119 - 1 CAUCACGAACAGCGCGAAGACUUCAAAAA-CGAUACAGCCAAGUGCCAAACACAAAUUAGUAGUAGGCAGUCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCCUGGCC .........(((.(((((((((((.....-.))....(((.(.(((.............))).).)))))))..........((((((((((......)))))))))).))))))))... ( -29.82) >DroYak_CAF1 27746 120 - 1 UAUCAUGAACAGCGCGAAGACUUCAAAAAGAGAUACAACCAAGUGCCUAACACAAAUUAGUAGUAGGCAGUCCGGGCACAAAGUGCUGGAAACUGAAGUUUCUUGCAUAUUUUCCUGGCC ..(((.(((.(..((((((((((((..................((((((..((......))..)))))).(((((.(((...))))))))...)))))))).))))...).))).))).. ( -30.50) >consensus CAUCACGAACAGCGCGAAGACUUCAAAAAGCGAUACAGCCAAGUGCCAAACACAAAUUAGUAGUAGGCAGGCCGAGCACAAAGUGCAGGAAAGUGAAGUUUCCUGCAUAUUUGCCUGGCC ..((.((.....)).))..........................((((.(..((......))..).))))(((((((((.((.((((((((((......)))))))))).))))))))))) (-27.69 = -28.62 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:16 2006