| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,480,206 – 4,480,309 |
| Length | 103 |
| Max. P | 0.727846 |

| Location | 4,480,206 – 4,480,309 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
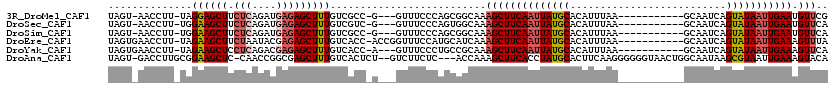
>3R_DroMel_CAF1 4480206 103 - 27905053 UAGU-AACCUU-UAGGAGCUUCUCAGAUGAGAGCUUUGUCGCC-G---GUUUCCCAGCGGCAAAGCUUCAAUUAUGCACAUUUAA-----------GCAAUCAGUAUAAUUGAAUGUUCG ....-......-...((((.((((....))))((((((((((.-(---(....)).))))))))))(((((((((((........-----------.......))))))))))).)))). ( -34.86) >DroSec_CAF1 33220 103 - 1 UAGU-AACCUU-UGGAAGCUUCUCAGAUGAGAGCUUUGUCGUC-G---GUUUCCCAGUGGCAAAGCUUCAAUUAUGCACAUUUAA-----------GCAAUCAGUAUAAUUGAAUGUUCA ....-......-..(((...((((....))))(((((((((..-(---(....))..)))))))))(((((((((((........-----------.......)))))))))))..))). ( -27.96) >DroSim_CAF1 34070 103 - 1 UAGU-AACCUU-UGGAAGCUUCUCAGAUGAGAGCUUUGUCGCC-G---GUUUCCCAGCGGCAAAGCUUCAAUUAUGCACAUUUAA-----------GCAAUCAGUAUAAUUGAAUGUUCA ....-......-..(((...((((....))))((((((((((.-(---(....)).))))))))))(((((((((((........-----------.......)))))))))))..))). ( -31.96) >DroEre_CAF1 34142 107 - 1 UAGUGAACCUU-UAGAAGCUUCUAAUACGAGAGCUUUGUCACC-ACCGGUUUCCAUGCAUCAAAGCUUCAAUUAUGCACAUUUAA-----------GCAAUCAGUAUAAUUGAAAGUUUA ..(..((((..-..((((((.((......))))))))......-...))))..)........(((((((((((((((........-----------.......))))))))).)))))). ( -21.58) >DroYak_CAF1 34832 104 - 1 UAGUGAACCUU-UAGAAGCUCCUCAGACGAGAGCUUUGUCACC-A---GUUUCCCUGCCGCAAAGCUUCAAUUAUGCACAUUUAA-----------GCAAUCAGUAUAAUUGAAAGUUCA ...(((((...-.........(((....))).(((((((...(-(---(.....)))..)))))))(((((((((((........-----------.......))))))))))).))))) ( -24.16) >DroAna_CAF1 34988 113 - 1 UAGU-GACCUUGCGGAAGCUC-CAACCGGCGAGCUUUGUCACUCU--GUCUUCUC---ACCAAAGCUUCACCUAUGCACUUCAAGGGGGGUAACUGGCAAUAAGCGUAAUUGAAAGUACA .(((-(((......(((((((-........)))))))))))))..--(((((.((---(.....((((..((..(((.(((....))).)))...))....)))).....)))))).)). ( -23.90) >consensus UAGU_AACCUU_UAGAAGCUUCUCAGAUGAGAGCUUUGUCACC_G___GUUUCCCAGCGGCAAAGCUUCAAUUAUGCACAUUUAA___________GCAAUCAGUAUAAUUGAAAGUUCA ..............((((((.(((....)))))))))..........................((((((((((((((..........................))))))))))).))).. (-15.72 = -16.47 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:34 2006