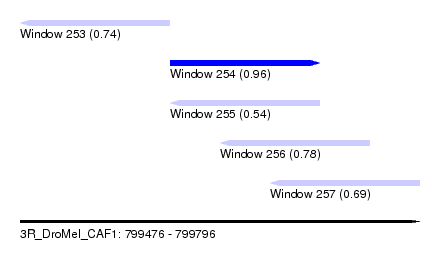
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 799,476 – 799,796 |
| Length | 320 |
| Max. P | 0.964799 |

| Location | 799,476 – 799,596 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 799476 120 - 27905053 UACAAGCGGGCAAAUGUUUAAAUACUUGAGUGCCGUACACGAGCAUAUUAACUUUUACUCAGCGCUCAGGGUUCGAAAUUGUCAACAAGACAAUUCACUUAUAUACAUAGAUCUGCACUU .....(((((...((((.((....(((((((((.(.....(((........))).....).)))))))))....(((.(((((.....)))))))).....)).))))...))))).... ( -28.60) >DroSec_CAF1 37497 120 - 1 UACAAGCGGGCAAAUGUUUAAAUACUUGAGUGCCGUACACGAGCAUAUUAACUUUUACUCAGCGCUCAGGGUUCGAAAUUGUCAACACGACAAUUCACUUACAUACAUAGAUCUGCACUU .....(((((...((((.......(((((((((.(.....(((........))).....).)))))))))....(((.(((((.....))))))))........))))...))))).... ( -28.00) >DroEre_CAF1 21190 110 - 1 UACGAGCGGGCAAAUGUUUAAAUACUUGAGUGCCGUACACGAGCAUAUUAACUUUUACUCAGCGCUCAGUGUUCGA--UUGUCAACACGACAAUUCACUU--------AGAUCUGCACUU .....((((((.........((((((.((((((.(.....(((........))).....).)))))))))))).((--(((((.....))))))).....--------.).))))).... ( -25.70) >DroYak_CAF1 27214 112 - 1 UACUAGCAGGCAAAUGUUUAAAUACUUGAGUGCCGUACACGAGCAUAUUAACUUUUGCUCAGCGCUCAGUGUUUGAAAGUGUCAACACGAAAAUUCACUU--------AGAUCUGUGCUU ....((((((((....((((((((((.((((((.(....)(((((..........))))).))))))))))))))))..)))).....((....))....--------.......)))). ( -31.30) >consensus UACAAGCGGGCAAAUGUUUAAAUACUUGAGUGCCGUACACGAGCAUAUUAACUUUUACUCAGCGCUCAGGGUUCGAAAUUGUCAACACGACAAUUCACUU________AGAUCUGCACUU .....((((((.............(((((((((.(.....(((........))).....).)))))))))....(((.(((((.....)))))))).............).))))).... (-21.17 = -22.05 + 0.88)

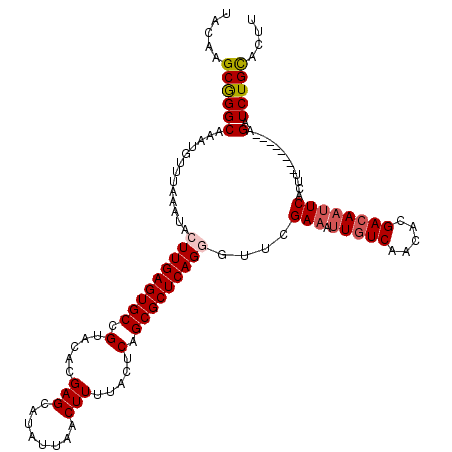
| Location | 799,596 – 799,716 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -44.80 |
| Energy contribution | -44.67 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 799596 120 + 27905053 AUUCGAGCCGAAUGCAAGUGCAGAUACGCGCAUCUGAGAGAAUGCCUUUGAAUUUCCGUAUCUGGGCGAUGCACAACGUCCUGGGAGAUGGCAUGUUCGGCAUGCGUGCUUGUUUGUAUC (((((...)))))....(((((((((((((((((((((...(((((.....((((((......(((((........)))))..))))))))))).))))).)))))))..))))))))). ( -41.00) >DroSec_CAF1 37617 120 + 1 AUUCGAGCCGCAUGCAAGUGCAGAUACGCGCAUCUGAGAGAAUGCCUUUGAAUCUCCGUAUCUGGGCGAUGCACAACGUCCUGGGAGAUGGCAUGUUCGGCAUGCGUGCUUGUUUGUAUC ...((((((((((((..((((......))))......(((.(((((.....((((((......(((((........)))))..))))))))))).))).))))))).)))))........ ( -46.80) >DroEre_CAF1 21300 120 + 1 AUUCGAGCCGCAUGCAAGUGCAGAUACGCGCAUCUGAGAGAAUGCCUUUGAAUCUCCGUAUCAGGGCGAUGUACAACGUCCUGGGAGAUGGCAUGUUCGGCAUGCGUGCUUGUUUGUAUC ...((((((((((((..((((......))))......(((.(((((.....((((((....(((((((........)))))))))))))))))).))).))))))).)))))........ ( -51.00) >DroYak_CAF1 27326 120 + 1 AUUCGAGCCGCAUGCAAGUGCAGAUACGCGCAUCUGAGAAAAUGCCUUUGAAUCUCCGUAUCUGGGCGAUGUACAACGUCCUGGGAGAUGGCAUGUUCGGCAUGCGUGCUUGUUUGUAUC ...((((((((((((..((((......))))......(((.(((((.....((((((......(((((........)))))..))))))))))).))).))))))).)))))........ ( -47.10) >consensus AUUCGAGCCGCAUGCAAGUGCAGAUACGCGCAUCUGAGAGAAUGCCUUUGAAUCUCCGUAUCUGGGCGAUGCACAACGUCCUGGGAGAUGGCAUGUUCGGCAUGCGUGCUUGUUUGUAUC ...((((((((((((..((((......))))......(((.(((((.....((((((......(((((........)))))..))))))))))).))).))))))).)))))........ (-44.80 = -44.67 + -0.12)

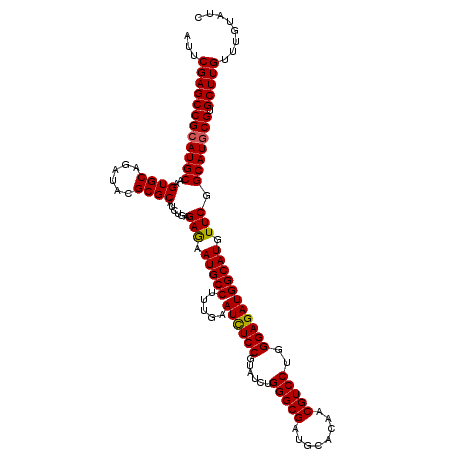
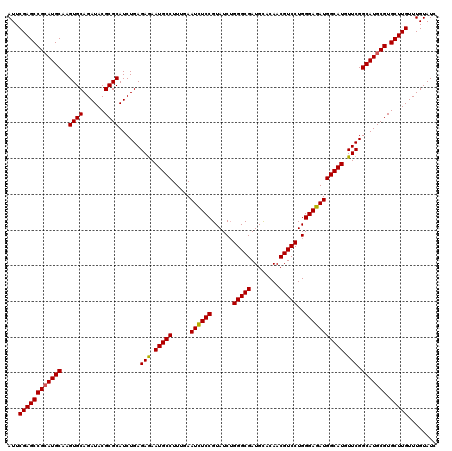
| Location | 799,596 – 799,716 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -33.35 |
| Energy contribution | -33.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
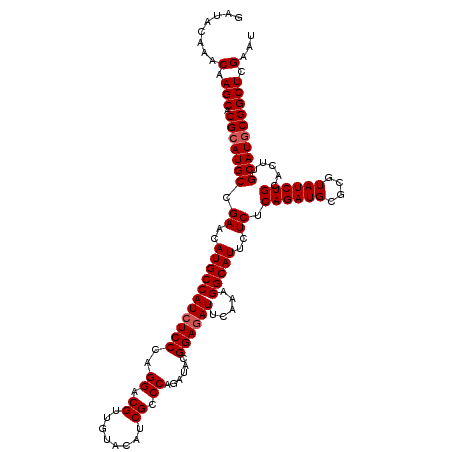
>3R_DroMel_CAF1 799596 120 - 27905053 GAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUGCAUCGCCCAGAUACGGAAAUUCAAAGGCAUUCUCUCAGAUGCGCGUAUCUGCACUUGCAUUCGGCUCGAAU ..............((...(((((..(((((........)).))).(((((.....(((((((((.....)....(((((......))))).))))))))....)))))))))).))... ( -29.90) >DroSec_CAF1 37617 120 - 1 GAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUGCAUCGCCCAGAUACGGAGAUUCAAAGGCAUUCUCUCAGAUGCGCGUAUCUGCACUUGCAUGCGGCUCGAAU ........(.(((.(((((((.((..(((((((((((..((.((........)).))......)))))).....)))))..)).((((((....)))))).....)))))))))).)... ( -36.20) >DroEre_CAF1 21300 120 - 1 GAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCUGAUACGGAGAUUCAAAGGCAUUCUCUCAGAUGCGCGUAUCUGCACUUGCAUGCGGCUCGAAU ........(.(((.(((((((.((..(((((((((((((((.((........)).))))....)))))).....)))))..)).((((((....)))))).....)))))))))).)... ( -41.10) >DroYak_CAF1 27326 120 - 1 GAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCAGAUACGGAGAUUCAAAGGCAUUUUCUCAGAUGCGCGUAUCUGCACUUGCAUGCGGCUCGAAU ........(.(((.(((((((.(((.(((((((((((..((.((........)).))......)))))).....))))).))).((((((....)))))).....)))))))))).)... ( -38.00) >consensus GAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCAGAUACGGAGAUUCAAAGGCAUUCUCUCAGAUGCGCGUAUCUGCACUUGCAUGCGGCUCGAAU ........(.(((.(((((((.((..(((((((((((..((.((........)).))......)))))).....)))))..)).((((((....)))))).....)))))))))).)... (-33.35 = -33.85 + 0.50)

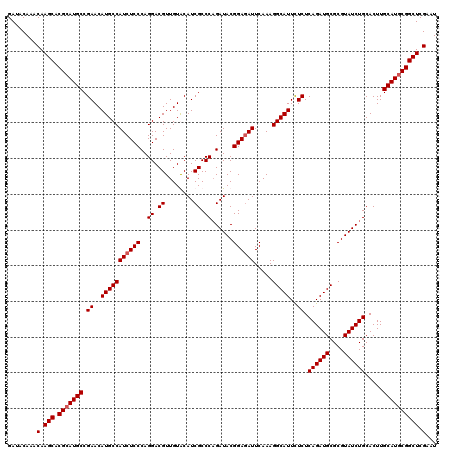
| Location | 799,636 – 799,756 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 799636 120 - 27905053 GAGCGCAGCGUCUGCGAAUGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUGCAUCGCCCAGAUACGGAAAUUCAAAGGCAUU (((((((((((((..(((((..........(((((.....)))))..........(((((.....)))))))).))...))))))))))).))(((..((.........))...)))... ( -35.40) >DroSec_CAF1 37657 120 - 1 GAGCGCAGCGUCUGCGAGCGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUGCAUCGCCCAGAUACGGAGAUUCAAAGGCAUU (((((((((((((((..((.....))....(((((.....)))))......))..(((((.....))))).........))))))))))).))(((..((.........))...)))... ( -38.00) >DroEre_CAF1 21340 120 - 1 GAGCGCAGCGUCUGCGAACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCUGAUACGGAGAUUCAAAGGCAUU ..(.(((((((((((.........))....(((((.....))))).....)).)))).))))....(((((((((((((((.((........)).))))....)))))).....))))). ( -34.10) >DroYak_CAF1 27366 120 - 1 GAGCGCAGCGUCUGCGGACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCAGAUACGGAGAUUCAAAGGCAUU (((..((((((((..(((.((.........(((((.....)))))..........(((((.....)))))..)))))..))))))))..).))(((..((.........))...)))... ( -32.60) >consensus GAGCGCAGCGUCUGCGAACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAGGACGUUGUACAUCGCCCAGAUACGGAGAUUCAAAGGCAUU (((((((((((((((.........))....(((((.....)))))..........(((((.....))))).........))))))))))).))(((..((.........))...)))... (-31.30 = -31.80 + 0.50)

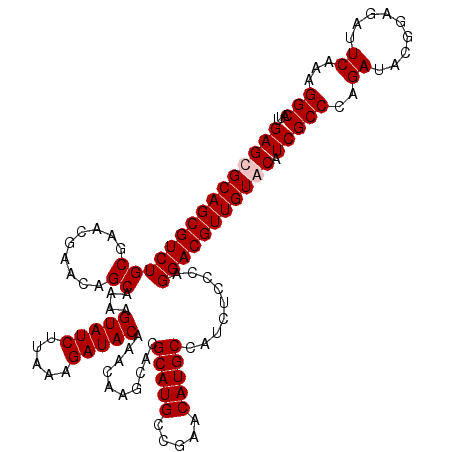
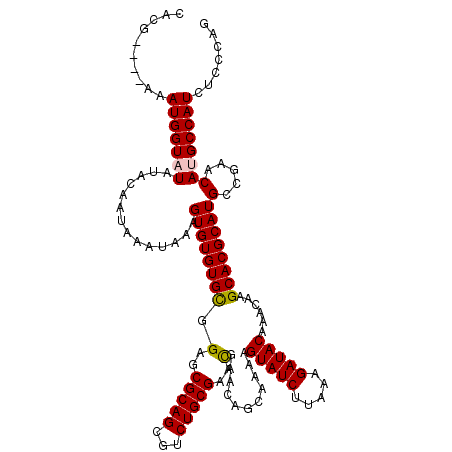
| Location | 799,676 – 799,796 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -25.31 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 799676 120 - 27905053 CACGAACGAAAUGGUUUAUACAAUAAAUAAAGUGUGUGUGGAGCGCAGCGUCUGCGAAUGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAG .......((.(((((((((.......)))).(((((((((...(((((...)))))..............(((((.....))))).......))))))))).......))))).)).... ( -26.80) >DroSec_CAF1 37697 120 - 1 CACGAACGAAAUGGUUUAUACAAUAAAUAAAGUGUGUGUGGAGCGCAGCGUCUGCGAGCGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAG .......((.(((((((((.......)))).((((((((((..(((((...)))))..)...........(((((.....))))).......))))))))).......))))).)).... ( -28.50) >DroEre_CAF1 21380 116 - 1 CACG----AAAUGGUAUAUACAAUAAAUAAAGUGUGUGCGGAGCGCAGCGUCUGCGAACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAG ...(----(.(((((((..............(((((((((...(((((...)))))..)...........(((((.....)))))......))))))))(.....)))))))).)).... ( -30.10) >DroYak_CAF1 27406 116 - 1 CACG----AAAUGGUAUAUACAAUAAAUAAAGUGUGUGCGGAGCGCAGCGUCUGCGGACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAG ...(----(.(((((((..............((((((((...((....((((....))))....))....(((((.....)))))......))))))))(.....)))))))).)).... ( -32.30) >consensus CACG____AAAUGGUAUAUACAAUAAAUAAAGUGUGUGCGGAGCGCAGCGUCUGCGAACGAACAGCAAAAGUAUCUUAAAGAUACAAACAAGCACGCAUGCCGAACAUGCCAUCUCCCAG ..........(((((((..............((((((((.(..(((((...)))))..)...........(((((.....)))))......))))))))(.....))))))))....... (-25.31 = -25.38 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:11 2006