
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,416,354 – 4,416,453 |
| Length | 99 |
| Max. P | 0.644665 |

| Location | 4,416,354 – 4,416,453 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -15.82 |
| Energy contribution | -17.05 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
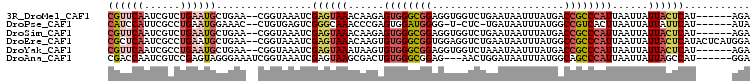
>3R_DroMel_CAF1 4416354 99 + 27905053 CGUUCAAUCGUCUGAAUGCUGAA--CGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAU------AGA ((((((..(((....))).))))--)).......((((((.....(((((((((..((((..........))))..))))))))).....))))))..------... ( -29.60) >DroPse_CAF1 25118 96 + 1 CAUCCAUUCGCCUGAAUGGAAAC--CUGUGAGUCGGGCAAACCCGAGUGGAUGGGG-U-CUC-UGAUAAUUUAUGGCCGUCACUUAAUUAUUAUUCAU------AUA ..(((((((....)))))))...--.(((((((.(((....)))((((((....((-(-(..-(((....))).)))).)))))).......))))))------).. ( -26.10) >DroSim_CAF1 19060 99 + 1 CGUUCAAUCGUCUGAAUGCUGAA--CGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAU------AGA ((((((..(((....))).))))--)).......((((((.....(((((((((..((((..........))))..))))))))).....))))))..------... ( -29.60) >DroEre_CAF1 17152 105 + 1 CGCUCAAUCGCCUGAAUGCUGAA--CGGUAAAUCGAGUAAACAAGUGUGGGCGGUGGAGGUCUGAAUAAUUUAUGGCCGCCCAUUAAUUAUUACUCAUACUCAUGGA ..........(((((.((((...--.))))....((((((......(((((((((.(((((.......)))).).)))))))))......))))))....))).)). ( -28.90) >DroYak_CAF1 17559 99 + 1 CGUUCAAUCGCCUGAAUGCUGAA--CGGUAAAUCGAGUAAAUAAGUGUGGGCGGAGGUGGUCUAAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAU------AGA ((((((...((......))))))--)).......((((((.(((..((((((((..((((..........))))..))))))))...)))))))))..------... ( -30.10) >DroAna_CAF1 22693 98 + 1 CGACCAAUCGUCCGAGUAGGGAAAUCGGUAAAUCGAGUAAGCGACUGUGGGCGGAG---AACUGGAUAAUUUAUGGCAGCCCAUUAAUUAUUAGCCAU------GGA ...(((.((((((((((.......((((....)))).......))).)))))))..---.....(((((((.((((....)))).))))))).....)------)). ( -23.94) >consensus CGUUCAAUCGCCUGAAUGCUGAA__CGGUAAAUCGAGUAAACAAGAGUGGGCGGAGGUGGUCUGAAUAAUUUAUGACCGCCCAUUAAUUAUUACUCAU______AGA ((((((......))))))................((((((......((((((((......................))))))))......))))))........... (-15.82 = -17.05 + 1.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:09 2006