| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,376,386 – 4,376,489 |
| Length | 103 |
| Max. P | 0.536599 |

| Location | 4,376,386 – 4,376,489 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.01 |
| Mean single sequence MFE | -39.97 |
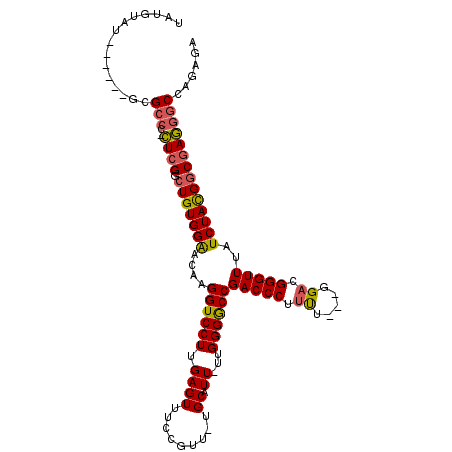
| Consensus MFE | -28.71 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
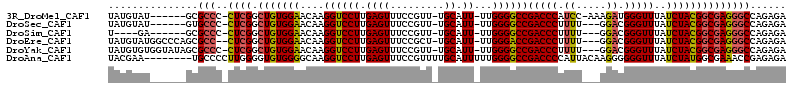
>3R_DroMel_CAF1 4376386 103 - 27905053 UAUGUAU------GCGCCC-CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGUU-UGCAUU-UUGGGGCCGACCCAUCC-AAAGAUGGGUUUAUCUACGGCGAGGGCCAGAGA ......(------(.((((-.(((.(((((((....((((((.((((.......-...)))-).))))))((((((((.-...))))))))..)))))))))))))))).... ( -42.90) >DroSec_CAF1 63452 101 - 1 UAUGUAU------GUGCCC-CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGUU-UGCAUU-UUGGGGCCGACCCUUUU---GGACGGGUUUAUCUACGGCGAGGGCCAGAGA ......(------.((.((-((((.(((((((....((((((.((((.......-...)))-).))))))(((((....---....)))))..))))))))))))).)).).. ( -38.60) >DroSim_CAF1 53827 97 - 1 U----GA------GCGCCC-CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGUU-UGCAUU-UUGGGGCCGACCCUUUU---GGACGGGUUUAUCUACGGCGAGGGCCAGAGA .----..------..(.((-((((.(((((((....((((((.((((.......-...)))-).))))))(((((....---....)))))..))))))))))))).)..... ( -37.50) >DroEre_CAF1 64759 106 - 1 UAUGUAUGGCCCAGCGCC--CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGCU-UGCAUU-UUGGGACCGACCCUUUU---GGACGGGUUUAUCUACGGCGAGGGCCAGAGA .............(.(((--((((.(((((((....((((((.((((.......-...)))-).))))))(((((....---....)))))..)))))))))))))))..... ( -41.30) >DroYak_CAF1 56061 107 - 1 UAUGUGUGGUAUAGCGCCC-CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGUU-UGCAUU-UUGGGGCCGACCCUUUU---GGACGGGUUUAUCUACGGCGAGGGCCAGAGA ...((((......))))((-((((.(((((((....((((((.((((.......-...)))-).))))))(((((....---....)))))..)))))))))))))....... ( -39.20) >DroAna_CAF1 61491 105 - 1 UACGAA--------UGCCCCUUGGGGUGUGGGGCAAGGUCCUUGAGUUUCCGUUUUGCAUUUUUGGGGCCGACCCCAUUACAAGGGGGGUUUAUCUAUGGCGAAACCGAGAGA ...(((--------(.(((((((....((((((...((((((.(((..............))).))))))..))))))..))))))).)))).(((.(((.....)))))).. ( -40.34) >consensus UAUGUAU______GCGCCC_CUCGGCUGUGGAACAAGGUCCUUGAGUUUCCGUU_UGCAUU_UUGGGGCCGACCCUUUU___GGACGGGUUUAUCUACGGCGAGGGCCAGAGA ...............(((..((((.(((((((....((((((.((((.........)).))...))))))(((((.((.....)).)))))..))))))))))))))...... (-28.71 = -28.72 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:57 2006