
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,335,556 – 4,335,653 |
| Length | 97 |
| Max. P | 0.808399 |

| Location | 4,335,556 – 4,335,653 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -20.43 |
| Energy contribution | -21.68 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
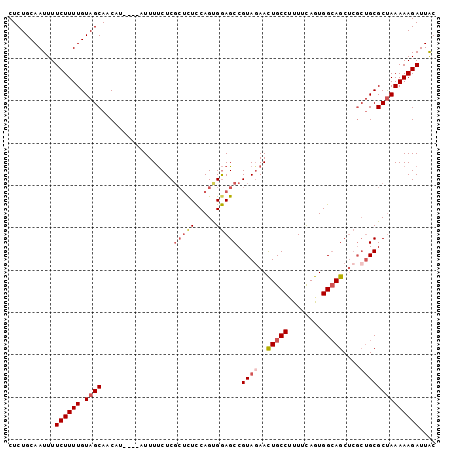
>3R_DroMel_CAF1 4335556 97 + 27905053 CUCUGCAAUUUUCUUUUGUAGCAACAU----AUUUUCUCGCUCUCCAGUGGAGCCGUAGAACUGCCUUUUUAGUGGCAGCUCGCUGCGCUAAAAAGAUUAC ...........((((((.((((.....----........(((((.....))))).((((..(((((........)))))....)))))))))))))).... ( -27.30) >DroSec_CAF1 22786 97 + 1 CUCUGCAAUUUUCUUUUGUAGCAACAU----AUUUUCUCGCUCUCCAGUGGAGCCGUAGAACUGCCUUUUCAGUGGGAGCUCGCUGCGCUAAAAAGAUUAC ...........(((((((((((((...----...))...((((.(((.(((((..(((....)))..))))).)))))))..)))))....)))))).... ( -22.80) >DroSim_CAF1 12135 97 + 1 CUCUGCAAUUUUCUUUUGUAGCAACAU----AUUUUCUCGCUUUCCAGUGGAGCCGUAGAACUGCCUUUUCAGUGGCAGCUCGCUGCGCUAAAAAGAUUAC ...........((((((.((((.....----....((.((((....))))))((.((.((.(((((........))))).)))).)))))))))))).... ( -25.30) >DroEre_CAF1 23025 97 + 1 CUCUGCAAUUUUCUUUUGUAGCAACAU----AUUUUCUCGCUCUCCAGCGGAGCCGUACAACUGCCUUUUCAGUGGCAGCUCGCUGCGCCAAAAAGAUUAC ...........(((((((((((.....----.......((((....))))((((.((...((((......)))).)).)))))))))....)))))).... ( -24.90) >DroYak_CAF1 14665 97 + 1 CUCUGCAAUUUUCUUUUGUAGCAACAU----AUUUUCUCGCUCUCCAGUGGAGCCGUAGAACUGCCUUUUCAGUGGCAGCUCGCUGCGCUAAAAAGAUUAC ...........((((((.((((.....----........(((((.....))))).((((..(((((........)))))....)))))))))))))).... ( -27.30) >DroAna_CAF1 10803 99 + 1 CUCUGCAAUUUUCUUUUGUAGCAACAUACAUAUUUCCUC--CCUUCAGUGAAGCCGUAGAAUUGCCUUUUCAAUGGCAGCAACGAGCGCUAAAAAGAUUGC ....((((...((((((.((((...........(((((.--.....)).)))(((((.(...((((........)))).).))).)))))))))))))))) ( -22.00) >consensus CUCUGCAAUUUUCUUUUGUAGCAACAU____AUUUUCUCGCUCUCCAGUGGAGCCGUAGAACUGCCUUUUCAGUGGCAGCUCGCUGCGCUAAAAAGAUUAC ...........((((((.((((.................(((((.....))))).((((..(((((........)))))....)))))))))))))).... (-20.43 = -21.68 + 1.25)


| Location | 4,335,556 – 4,335,653 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4335556 97 - 27905053 GUAAUCUUUUUAGCGCAGCGAGCUGCCACUAAAAAGGCAGUUCUACGGCUCCACUGGAGAGCGAGAAAAU----AUGUUGCUACAAAAGAAAAUUGCAGAG ((((((((((....(((((((((((((........))))))))..((.((((...))))..)).......----..)))))...))))))...)))).... ( -30.50) >DroSec_CAF1 22786 97 - 1 GUAAUCUUUUUAGCGCAGCGAGCUCCCACUGAAAAGGCAGUUCUACGGCUCCACUGGAGAGCGAGAAAAU----AUGUUGCUACAAAAGAAAAUUGCAGAG ((((((((((....((((((((((.((........)).)))))..((.((((...))))..)).......----..)))))...))))))...)))).... ( -23.90) >DroSim_CAF1 12135 97 - 1 GUAAUCUUUUUAGCGCAGCGAGCUGCCACUGAAAAGGCAGUUCUACGGCUCCACUGGAAAGCGAGAAAAU----AUGUUGCUACAAAAGAAAAUUGCAGAG ((((((((((....(((((((((((((........)))))))).....(((..((....)).))).....----..)))))...))))))...)))).... ( -28.20) >DroEre_CAF1 23025 97 - 1 GUAAUCUUUUUGGCGCAGCGAGCUGCCACUGAAAAGGCAGUUGUACGGCUCCGCUGGAGAGCGAGAAAAU----AUGUUGCUACAAAAGAAAAUUGCAGAG (((((.((((((..(((((((((((((........)))))))...((.((((...))))..)).......----.))))))..))))))...))))).... ( -29.90) >DroYak_CAF1 14665 97 - 1 GUAAUCUUUUUAGCGCAGCGAGCUGCCACUGAAAAGGCAGUUCUACGGCUCCACUGGAGAGCGAGAAAAU----AUGUUGCUACAAAAGAAAAUUGCAGAG ((((((((((....(((((((((((((........))))))))..((.((((...))))..)).......----..)))))...))))))...)))).... ( -30.50) >DroAna_CAF1 10803 99 - 1 GCAAUCUUUUUAGCGCUCGUUGCUGCCAUUGAAAAGGCAAUUCUACGGCUUCACUGAAGG--GAGGAAAUAUGUAUGUUGCUACAAAAGAAAAUUGCAGAG ((((((((((((((((.(((...((((........)))).....)))))(((.((.....--.)))))...........)))).))))))...)))).... ( -22.40) >consensus GUAAUCUUUUUAGCGCAGCGAGCUGCCACUGAAAAGGCAGUUCUACGGCUCCACUGGAGAGCGAGAAAAU____AUGUUGCUACAAAAGAAAAUUGCAGAG (((((((((((((((((..((((((((........))))))))..((.((((...))))..))............)).))))).))))))...)))).... (-23.30 = -24.08 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:43 2006