
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,326,783 – 4,326,890 |
| Length | 107 |
| Max. P | 0.806347 |

| Location | 4,326,783 – 4,326,890 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4326783 107 - 27905053 CAGUUGGUGUUGCAAUUAAAAGUGGAAUUCUUUUUCAUU----------AUUGCUGUUUGUCGGAAAACAGGCAAAUAUUAAUAAAGCAAACGCGCUCACAUGC--CCC-CAACCAUCCA ..(((((((((((.........(((((.....)))))((----------((((.((((((((........)))))))).)))))).)).)))))((......))--...-))))...... ( -24.60) >DroPse_CAF1 5843 117 - 1 CAGUUGAUGUUGCAAUUAAGAGUGGAAUUCUUUUUCAUUUGGUGCUUGUGUUUCUGUUUGCCCAAAAACAGGCAAAUAUUAAUAAAGCAAGCUCCCACAGAGAGAUCUCUC-UCCCUC-- .(((((......)))))..((((((((.....))))))))((.(((((((((..((((((((........))))))))..)))...)))))).))....(((((....)))-))....-- ( -33.30) >DroSec_CAF1 14119 106 - 1 CAGUUGGUGUUGCAAUUAAGAGUGGAAUUCUUUUUCAUU----------AUUGCUGUUUGUCGGAAAACAGGCAAAUAUUAAUAAAGCAAACGCGCACAUAUAC--GCC-C-ACCAUCCA ....(((((((((.........(((((.....)))))((----------((((.((((((((........)))))))).)))))).))))..(((........)--)).-)-)))).... ( -25.50) >DroEre_CAF1 14240 106 - 1 CAGUUGGUGUUGCAAUUAAGAGUGGAAUUCUUUUCCAUU----------AUUGCUGUUUGUCGGAAAACAGGCAAAUAUUAAUAAAGCAAAUGCGCACACAUAC--CCC-C-ACCAUCCA ....(((((((((.........(((((.....)))))((----------((((.((((((((........)))))))).)))))).))))(((......)))..--...-)-)))).... ( -25.40) >DroAna_CAF1 1881 100 - 1 CGGUUGGCGUUGCAAUUACGAGAGGAAUUCUUUUUCAUU----------AUUGCUGC------AAAAACAGGCAAAUAUUAAUAAAACACA-GCCCACACGCAU--CCCGC-UCCGCCCA (((...((((.(((((...((((((....))))))....----------)))))...------.......(((..................-)))...))))..--.))).-........ ( -18.27) >DroPer_CAF1 2354 118 - 1 CAGUUGAUGUUGCAAUUAAGAGUGGAAUUCUUUUUCAUUUGGUGCUUGUGUUUCUGUUUGCCCAAAAACAGGCAAAUAUUAAUAAAGCAAGCUCCCACAGAGAGAUCUCCC-UCCCUCA- .(((((......)))))..(((.(((..(((((((.....((.(((((((((..((((((((........))))))))..)))...)))))).))...)))))))..))))-)).....- ( -34.40) >consensus CAGUUGGUGUUGCAAUUAAGAGUGGAAUUCUUUUUCAUU__________AUUGCUGUUUGUCGAAAAACAGGCAAAUAUUAAUAAAGCAAACGCCCACACAUAC__CCC_C_ACCAUCCA ..(((((((((((.......(((((((.....)))))))..............((((((......)))))))).)))))))))..................................... (-14.99 = -15.47 + 0.47)

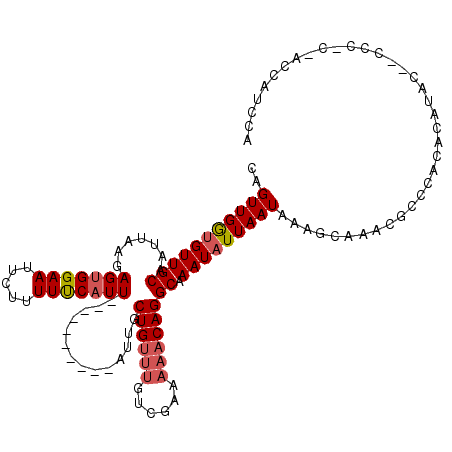
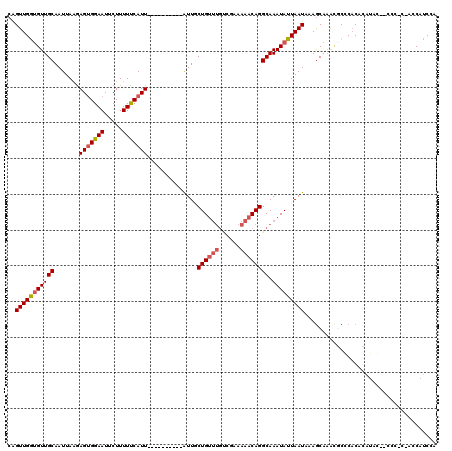
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:37 2006