
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,315,978 – 4,316,278 |
| Length | 300 |
| Max. P | 0.993748 |

| Location | 4,315,978 – 4,316,082 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.47 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -26.72 |
| Energy contribution | -26.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
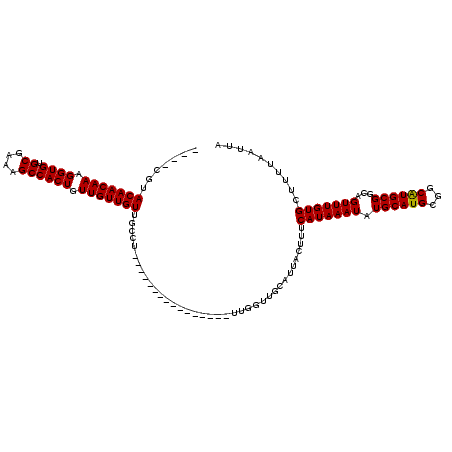
>3R_DroMel_CAF1 4315978 104 + 27905053 AAUACGUACAACAAAGGUGUGCGAAAGCCACUGUUGUUGUUGCCU----------------UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUA .....(((((((((.((((.((....)))))).))))).(((((.----------------...(((((((.................)))))))....)))))...))))......... ( -30.63) >DroSec_CAF1 3368 116 + 1 ----CGUACAACAAAGGUGUGCGAAAGCCACUGUUGUUGUUGCCUUUGCAAAGGCAAAGCUUUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUA ----.(((((((((.((((.((....)))))).))))).((((((((((....)))))((....(((((((.................)))))))..)))))))...))))......... ( -36.33) >DroEre_CAF1 3378 100 + 1 ----CGUACAACAAAGGUGUGCGAAGGCCACUGUUGUUGUUGGCU----------------UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCGUGCGGCAGUUUGUGCUUUUAAUUA ----((.(((((((.((((.((....)))))).)))))))))((.----------------......))........(((((((.((((((...))))))...))))))).......... ( -27.10) >consensus ____CGUACAACAAAGGUGUGCGAAAGCCACUGUUGUUGUUGCCU________________UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUA .......(((((((.((((.((....)))))).))))))).....................................(((((((.((((((...))))))...))))))).......... (-26.72 = -26.50 + -0.22)

| Location | 4,315,978 – 4,316,082 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.47 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4315978 104 - 27905053 UAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA----------------AGGCAACAACAACAGUGGCUUUCGCACACCUUUGUUGUACGUAUU .........((......((((((.....)).(((((...(((....)))))))).....----------------.))))(((((((..(((((....)).)))..)))))))..))... ( -25.50) >DroSec_CAF1 3368 116 - 1 UAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAAAGCUUUGCCUUUGCAAAGGCAACAACAACAGUGGCUUUCGCACACCUUUGUUGUACG---- ......(((((......(((.(((((...)))))....((((....)))).)))......)))))(((((....))))).(((((((..(((((....)).)))..)))))))...---- ( -30.60) >DroEre_CAF1 3378 100 - 1 UAAUUAAAAGCACAAACUGCCGCACGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA----------------AGCCAACAACAACAGUGGCCUUCGCACACCUUUGUUGUACG---- .........((......(((.(....).)))(((((...(((....)))))))).....----------------.))..(((((((..(((((....)).)))..)))))))...---- ( -20.60) >consensus UAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA________________AGGCAACAACAACAGUGGCUUUCGCACACCUUUGUUGUACG____ .................((((((.....)).(((((...(((....))))))))......................))))(((((((..(((((....)).)))..)))))))....... (-22.37 = -22.70 + 0.33)



| Location | 4,316,018 – 4,316,122 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
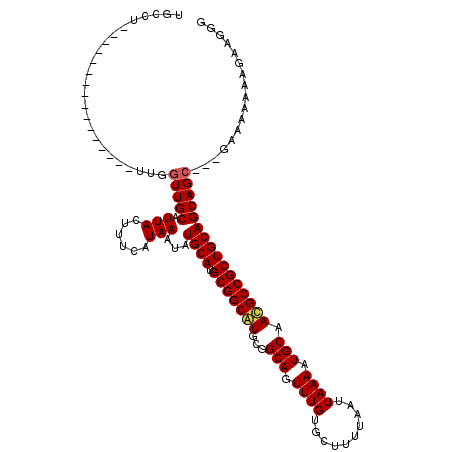
>3R_DroMel_CAF1 4316018 104 + 27905053 UGCCU----------------UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAAUGCCGCUGCAGCAGCAGGGAAAAAAAGAAGCG ..(((----------------...(((((.(((......)))...((((.(((((((...(((.((((..........)))).))).)))))))))))))))).)))............. ( -30.00) >DroSec_CAF1 3404 115 + 1 UGCCUUUGCAAAGGCAAAGCUUUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAG-----AAAAAAAGAAGGG ((((((....))))))...((((...(((((.............)))))(((.(((.(((((.(((.((..(((.....)))..))))))))))))).))).-----.......)))).. ( -31.92) >DroEre_CAF1 3414 101 + 1 UGGCU----------------UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCGUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAGC---GGAAAAAAGAAGGG ....(----------------((.(((((.(((......)))...((((.(((((((...(((.((((..........)))).))).))))))))))))))))---.))).......... ( -30.60) >consensus UGCCU________________UUGGUUGCAUUACUUUCAUAAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAGC___GAAAAAAAGAAGGG ........................(((((.(((......)))...((((.(((((((...(((.((((..........)))).))).))))))))))))))))................. (-24.12 = -24.57 + 0.45)

| Location | 4,316,018 – 4,316,122 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4316018 104 - 27905053 CGCUUCUUUUUUUCCCUGCUGCUGCAGCGGCAUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA----------------AGGCA .((..(((((......((((((.(((((((((.(((.((((....))))))).....)))))).))).))).))).......)))))....))......----------------..... ( -28.32) >DroSec_CAF1 3404 115 - 1 CCCUUCUUUUUUU-----CUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAAAGCUUUGCCUUUGCAAAGGCA .............-----.(((.(((((((((((((.((((....)))))))...)).))))).))).)))(((((...(((....))))))))..........((((((....)))))) ( -30.80) >DroEre_CAF1 3414 101 - 1 CCCUUCUUUUUUCC---GCUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCACGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA----------------AGCCA ..............---..((((((((((((((.((.((((....))))(((.....))).))))))))).))))...((((....)))).))).....----------------..... ( -25.20) >consensus CCCUUCUUUUUUUC___GCUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUUAUGAAAGUAAUGCAACCAA________________AGGCA ...................((((((((((((((.((.((((....))))(((.....))).))))))))).))))...((((....)))).))).......................... (-24.58 = -24.13 + -0.44)



| Location | 4,316,042 – 4,316,160 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -23.90 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4316042 118 + 27905053 AAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAAUGCCGCUGCAGCAGCAGGGAAAAAAAGAAGCGAAACUACAAAAAAAAAA--AAACGGAACUGCACACAUAUU .((((((..(((.(((.((((((.....(((((((....)))).))).))))))))).)))((((................................--........))))...)))))) ( -28.80) >DroSec_CAF1 3444 112 + 1 AAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAG-----AAAAAAAGAAGGGAAACUACAAGAAA-AAA--AAGCGGAACUGCACACAUAUU .((((((..(((.(((.(((((.(((.((..(((.....)))..))))))))))))).))).-----..........((....))........-...--..(((....)))...)))))) ( -27.20) >DroEre_CAF1 3438 117 + 1 AAAUAUGCAUGCGGCGUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAGC---GGAAAAAAGAAGGGAAACUACAAAAAAAAAAUAAGGCGGAACUGCACACAUAUU .....((((((...))))))(((((((.(((((((....)))).))).((((.((((....))---)).........((....))...............)))))))))))......... ( -30.90) >consensus AAAUAUGCAUGCGGCAUGCGGCAGUUUGUGCUUUUAAUUAAAAUGCAACGCCGCUGCAGCAGC___GAAAAAAAGAAGGGAAACUACAAAAAAAAAA__AAGCGGAACUGCACACAUAUU .((((((..((((((.((((((.((...(((((((....)))).)))..)).)))))))).................((....)).......................))))..)))))) (-23.90 = -24.23 + 0.33)


| Location | 4,316,042 – 4,316,160 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -29.00 |
| Energy contribution | -28.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4316042 118 - 27905053 AAUAUGUGUGCAGUUCCGUUU--UUUUUUUUUUGUAGUUUCGCUUCUUUUUUUCCCUGCUGCUGCAGCGGCAUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUU (((((((((((..........--.........((((((...((..............)).))))))((((((.(((.((((....))))))).....)))))).....))))))))))). ( -31.64) >DroSec_CAF1 3444 112 - 1 AAUAUGUGUGCAGUUCCGCUU--UUU-UUUCUUGUAGUUUCCCUUCUUUUUUU-----CUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUU (((((((((((..........--...-.....((((((...............-----..))))))((((((((((.((((....)))))))...)).))))).....))))))))))). ( -29.43) >DroEre_CAF1 3438 117 - 1 AAUAUGUGUGCAGUUCCGCCUUAUUUUUUUUUUGUAGUUUCCCUUCUUUUUUCC---GCUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCACGCCGCAUGCAUAUUU (((((((((((.((..................((((((................---...))))))((((((((((.((((....)))))))...)).)))))..)).))))))))))). ( -29.31) >consensus AAUAUGUGUGCAGUUCCGCUU__UUUUUUUUUUGUAGUUUCCCUUCUUUUUUUC___GCUGCUGCAGCGGCGUUGCAUUUUAAUUAAAAGCACAAACUGCCGCAUGCCGCAUGCAUAUUU (((((((((((.....................((((((......................))))))((((((.(((.((((....))))))).....)))))).....))))))))))). (-29.00 = -28.78 + -0.22)


| Location | 4,316,160 – 4,316,278 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4316160 118 - 27905053 CAUCGCCGCACUCUAUGUAAUUGAAAUAUCCUGUCCA--AGGAUAGCGGCAAAAGUGGAGGCGAAAAUAGAAAAUGGGCCACGAGCUCAAGGAAUUCAAUCAAUUGCAAGGCGAACUUCG ..(((((((.((((((....(((...((((((.....--))))))....)))..))))))))............(((((.....)))))....................)))))...... ( -32.90) >DroSec_CAF1 3556 120 - 1 CAUCGUCGCACUCUACGUAAUUGAAAUAUGCUGUCCAAAAGGAUAGCGGCAGAAGUGGAGGCGAAAAUAGAAAGUGGGCCACGAGCUCAAGGAAUUCAAUCAAUUGCAAGGCGAACUUCA ..(((((((.((((((....(((.....((((((((....)))))))).)))..)))))))).......(((..(((((.....))))).....)))............)))))...... ( -35.60) >DroEre_CAF1 3555 118 - 1 CAUCGUCGCACUCUAUGUAAUUGAAAUAUGCUGUCCA--AGGAUAAGGGCAAAAAAGGAGGCGAAUGUAGAAAAUGGGCCACAAGCUCAAGGAAUUCAAUCAAUUGCAAGGCGAACUUCA .....((((.((...(((((((((.......(((((.--.......)))))...........((((........(((((.....)))))....))))..)))))))))))))))...... ( -26.70) >consensus CAUCGUCGCACUCUAUGUAAUUGAAAUAUGCUGUCCA__AGGAUAGCGGCAAAAGUGGAGGCGAAAAUAGAAAAUGGGCCACGAGCUCAAGGAAUUCAAUCAAUUGCAAGGCGAACUUCA ..(((((((.((((((....(((.......((((((....))))))...)))..))))))))............(((((.....)))))....................)))))...... (-27.78 = -27.57 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:09 2006