
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,298,116 – 4,298,216 |
| Length | 100 |
| Max. P | 0.639187 |

| Location | 4,298,116 – 4,298,216 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -19.09 |
| Energy contribution | -18.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4298116 100 + 27905053 UACAAUC------GAAUUCCAUCCU-------------ACCACGCAGCCGCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC-UGGGUU .......------............-------------.(((.((((..((((.((...(((((((........))))).)).)))))).....((((.....))))..))))-)))... ( -29.90) >DroSec_CAF1 99342 100 + 1 UACAAUC------CAAUCCCAUCCU-------------ACCACGCAGCCUCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC-UCGGUU .......------............-------------(((..((((....(((((...(((((((........))))).)).)))))......((((.....))))..))))-..))). ( -27.10) >DroSim_CAF1 99963 100 + 1 UACAAUC------CAAUCCCAUCCU-------------ACCACGCAGCCGCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC-UCGGUU .......------............-------------(((..((((..((((.((...(((((((........))))).)).)))))).....((((.....))))..))))-..))). ( -28.00) >DroEre_CAF1 102632 105 + 1 UACAAGCCCAUACCAAUACCAUCCU-------------ACCACUCAGCCGCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC-U-GGUU .........................-------------.....(((((.((((.((...(((((((........))))).)).)))))).....((((.....))))....))-)-)).. ( -26.40) >DroWil_CAF1 108280 113 + 1 UAUCCUC------CAAUCCCCCACCCCCCCCAUCGCUUAUAUAUCAGCCUUCCAUCGUCAUGUCGCUAAUUAAAGUGGGCGUGAAAAGCAAACAGAGUGC-AAAUUUAUCGUCAUUGGCU .......------................................((((......((((((((((((......))).)))))))...(((.......)))-........)).....)))) ( -16.30) >DroYak_CAF1 102858 100 + 1 CACAAUC------CAAUCCUAUACU-------------ACCUCUCAACCGCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC-UCGGCU .......------............-------------...........(((((((...(((((((........))))).)).))))(((....((((.....))))...)))-..))). ( -24.40) >consensus UACAAUC______CAAUCCCAUCCU_____________ACCACGCAGCCGCCCCGCACAGCCCCGCUAAUUAAUGUGGGCGUGGCGGGCAAACAAAGUGCUAAAUUUAUCUGC_UCGGUU .............................................((((..(((((...(((((((........))))).)).)))))......((((.....)))).........)))) (-19.09 = -18.98 + -0.11)

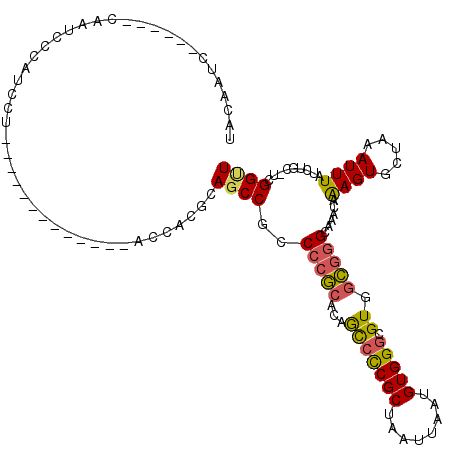
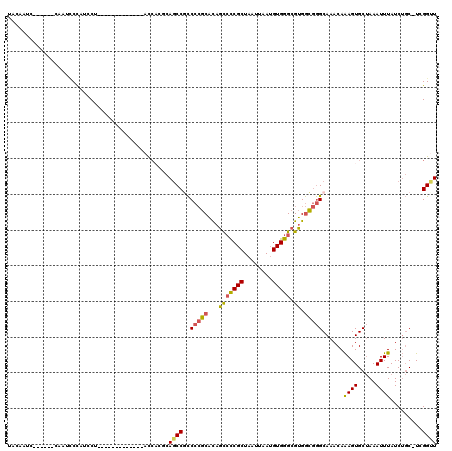
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:49 2006