
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,288,100 – 4,288,220 |
| Length | 120 |
| Max. P | 0.991430 |

| Location | 4,288,100 – 4,288,207 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977125 |
| Prediction | RNA |
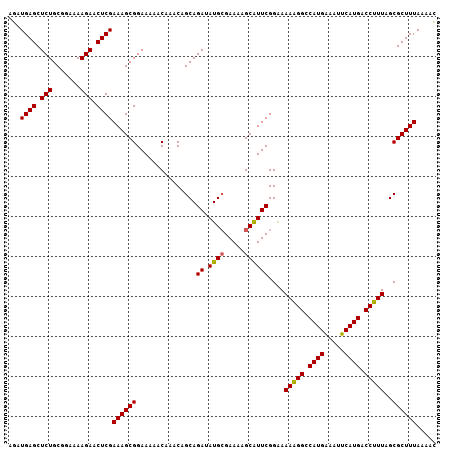
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4288100 107 + 27905053 AGAUGAGCUCUGCGGAAAAGAACUCGAAAGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGAAAAGGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAAU ...((((.(((.......))).))))((((((...............((.((((.....))))))...(((((..((((.....)))).)))))..))))))..... ( -26.10) >DroSec_CAF1 89538 107 + 1 UGAUGAGCUCUGCGGAAAAGAACUCGAAAGCGGAAAAACAAACAGCAGAUAUGGGAAAAGCAUUCGGAAAAAGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAAC ...((((.(((.......))).))))((((((...............((.(((.......)))))....(((((.((((.....)))).)))))..))))))..... ( -21.80) >DroSim_CAF1 88994 107 + 1 AGAUGAGCUCUGCGGAAAAGAACUCGAAAGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGGAAAAGGCCAUGGAAUUCAUGACCUUUAGCGCUUUAAAAC ...((((.(((.......))).))))((((((...............((.((((.....))))))....(((((.((((.....)))).)))))..))))))..... ( -25.40) >DroEre_CAF1 92572 106 + 1 AGAUGAGCUCUGCG-AAAAGAACUCGAAAGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCGUUCGGAAAAAGGCCAUGAAAUUCAUGGCCUUUAGCGCUUUAAAAU ...((((.(((...-...))).))))((((((...............((.((((.....))))))....((((((((((.....))))))))))..))))))..... ( -33.50) >consensus AGAUGAGCUCUGCGGAAAAGAACUCGAAAGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGAAAAAGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAAC ...((((.(((.......))).))))((((((...............((.((((.....))))))....(((((.((((.....)))).)))))..))))))..... (-24.62 = -24.50 + -0.12)

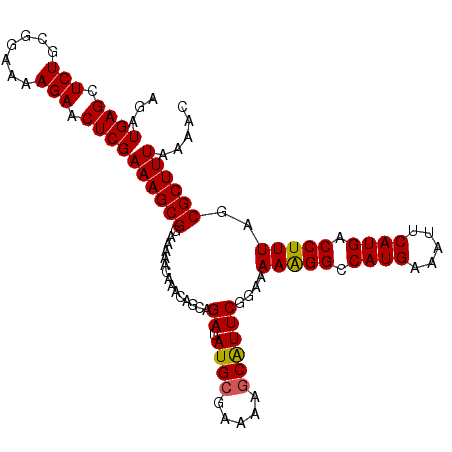
| Location | 4,288,100 – 4,288,207 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -29.17 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4288100 107 - 27905053 AUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCCUUUUCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCUUUCGAGUUCUUUUCCGCAGAGCUCAUCU .....((((((..((((((((((.....))))))).......((((((.....)))).))..........)))..)))))).(((((((.......))))))).... ( -28.00) >DroSec_CAF1 89538 107 - 1 GUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCUUUUUCCGAAUGCUUUUCCCAUAUCUGCUGUUUGUUUUUCCGCUUUCGAGUUCUUUUCCGCAGAGCUCAUCA .....((((((..((((((((((.....))))))))))...(((((((.............)).)))))......)))))).(((((((.......))))))).... ( -28.82) >DroSim_CAF1 88994 107 - 1 GUUUUAAAGCGCUAAAGGUCAUGAAUUCCAUGGCCUUUUCCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCUUUCGAGUUCUUUUCCGCAGAGCUCAUCU .....((((((..((((((((((.....))))))))))....((((((.....)))).))...............)))))).(((((((.......))))))).... ( -29.50) >DroEre_CAF1 92572 106 - 1 AUUUUAAAGCGCUAAAGGCCAUGAAUUUCAUGGCCUUUUUCCGAACGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCUUUCGAGUUCUUUU-CGCAGAGCUCAUCU .....((((((..((((((((((.....))))))))))...(((((((.............)).)))))......)))))).(((((((...-...))))))).... ( -33.82) >consensus AUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCUUUUUCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCUUUCGAGUUCUUUUCCGCAGAGCUCAUCU .....((((((..((((((((((.....))))))))))...(((((((.............)).)))))......)))))).(((((((.......))))))).... (-29.17 = -29.04 + -0.13)

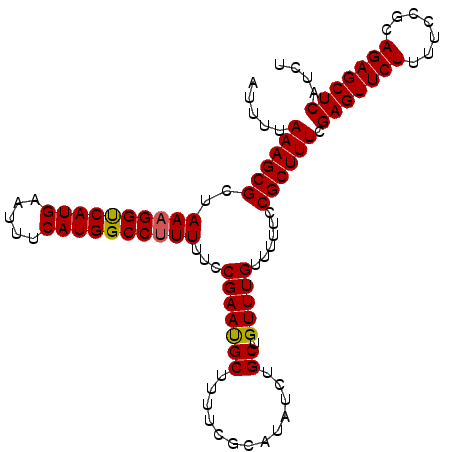

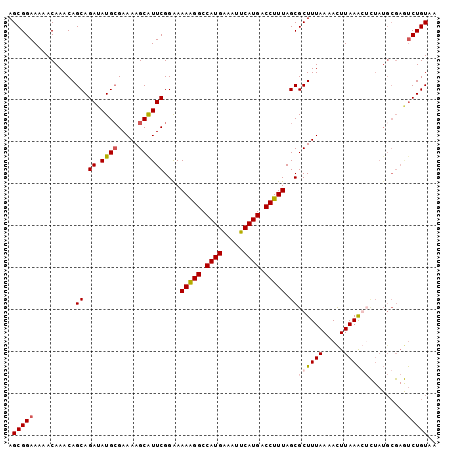
| Location | 4,288,128 – 4,288,220 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -17.11 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4288128 92 + 27905053 AGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGAAAAGGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAAUUUAAGC----------CCUGUAA .((((...........((.((.((((.....))))))...(((((..((((.....)))).))))).))((((........))))----------.)))).. ( -22.10) >DroSec_CAF1 89566 102 + 1 AGCGGAAAAACAAACAGCAGAUAUGGGAAAAGCAUUCGGAAAAAGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAACUUAAAGUCUAUGCGAGUCUGUAA .((.............))...((((((....((((...(..(((((.((((.....)))).)))))..)((((((.....))))))..))))...)))))). ( -19.82) >DroSim_CAF1 89022 102 + 1 AGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGGAAAAGGCCAUGGAAUUCAUGACCUUUAGCGCUUUAAAACUUAAAGUCUAUGCGAGUCUGUAA ................((((((((((.....))))(((.(.(((((.((((.....)))).)))))((.((((((.....)))))))).).))))))))).. ( -23.00) >DroEre_CAF1 92599 102 + 1 AGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCGUUCGGAAAAAGGCCAUGAAAUUCAUGGCCUUUAGCGCUUUAAAAUUUAAACAUUAUGCGAAUCUGUUA ...............(((((((.((((.((((((((.....((((((((((.....))))))))))))))))))..(((......))).)))).))))))). ( -33.00) >consensus AGCGGAAAAACAAACAGCAGAUAUGCGAAAAGCAUUCGGAAAAAGGCCAUGAAAUUCAUGACCUUUAGCGCUUUAAAACUUAAACUCUAUGCGAGUCUGUAA .(((((..........((.((.((((.....))))))....(((((.((((.....)))).))))).))((((((.....)))))).........))))).. (-17.11 = -18.05 + 0.94)

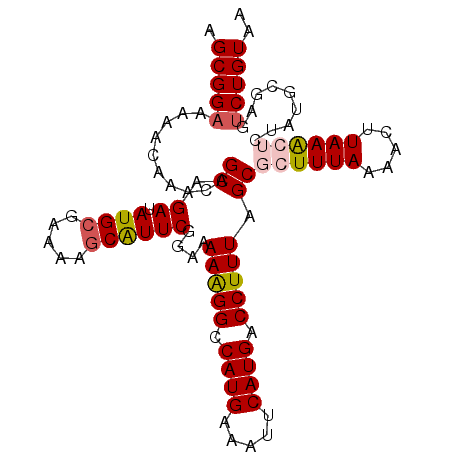
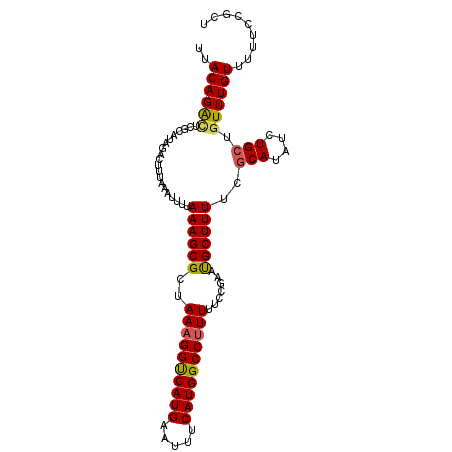
| Location | 4,288,128 – 4,288,220 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -19.49 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4288128 92 - 27905053 UUACAGG----------GCUUAAAUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCCUUUUCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCU .....((----------(...((((.....((((..((((((((((.....))))).)))))..)((((((.....)))).)).)))....)))).)))... ( -20.20) >DroSec_CAF1 89566 102 - 1 UUACAGACUCGCAUAGACUUUAAGUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCUUUUUCCGAAUGCUUUUCCCAUAUCUGCUGUUUGUUUUUCCGCU ..((((((..(((.((((.....)))).((((((..((((((((((.....)))))))))).......)))))).........))).))))))......... ( -23.50) >DroSim_CAF1 89022 102 - 1 UUACAGACUCGCAUAGACUUUAAGUUUUAAAGCGCUAAAGGUCAUGAAUUCCAUGGCCUUUUCCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCU ..((((((..((((...(((((.....)))))((..((((((((((.....))))))))))...)).)))).....(((....))).))))))......... ( -25.10) >DroEre_CAF1 92599 102 - 1 UAACAGAUUCGCAUAAUGUUUAAAUUUUAAAGCGCUAAAGGCCAUGAAUUUCAUGGCCUUUUUCCGAACGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCU .(((((((..(((..((((.........((((((..((((((((((.....)))))))))).......))))))..))))...))).)))))))........ ( -27.50) >consensus UUACAGACUCGCAUAGACUUUAAAUUUUAAAGCGCUAAAGGUCAUGAAUUUCAUGGCCUUUUUCCGAAUGCUUUUCGCAUAUCUGCUGUUUGUUUUUCCGCU ..((((((....................((((((..((((((((((.....)))))))))).......))))))..(((....))).))))))......... (-19.49 = -19.55 + 0.06)

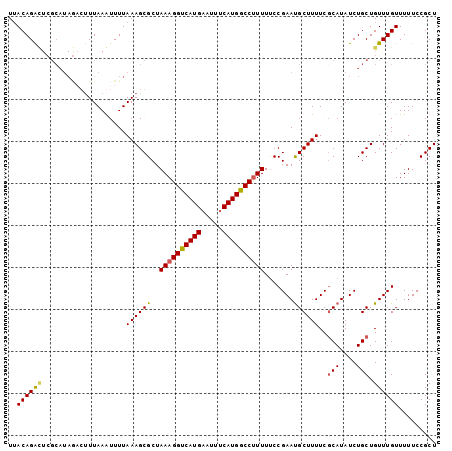
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:42 2006