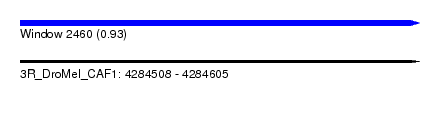
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,284,508 – 4,284,605 |
| Length | 97 |
| Max. P | 0.928429 |

| Location | 4,284,508 – 4,284,605 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.81 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
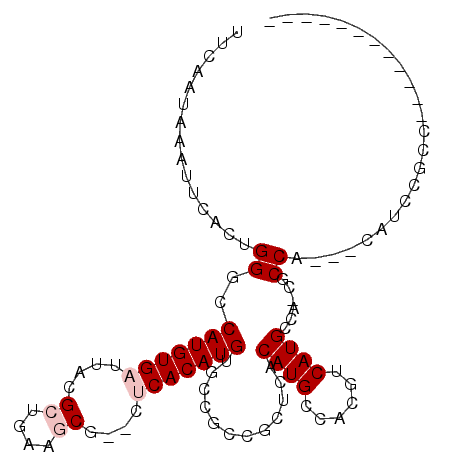
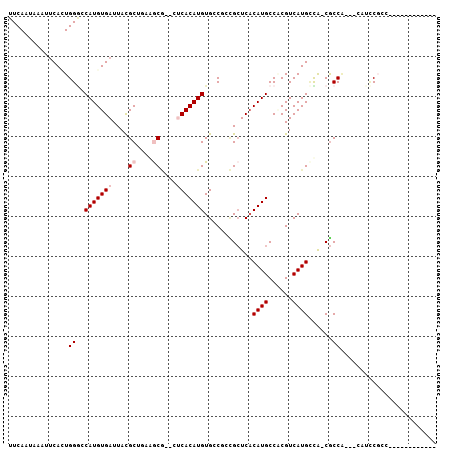
>3R_DroMel_CAF1 4284508 97 + 27905053 UUCAAUAAUUUCACUGGGCCAUGUGAUUACGCUGGCGCG--CUCACAUGUGCCUCCGCUCACAUGCCUCGUCAUGCCA-CGCCAUCGCAUCCGCCAUUU-------G ................(((.(((((((..((.(((((((--(......))))....((......))........))))-))..)))))))..)))....-------. ( -26.60) >DroVir_CAF1 125930 90 + 1 UUCAAUAAAUUCAGUGGAGCAUGUGAUUACGCAAAAACACAUACACAUGUGGCGGUGCUCACAUGCCACGUCAUGUUGCCGCCG---UCUUCU-------------- ..(((((......(((..((((((((....(((....(((((....)))))....))))))))))))))....)))))......---......-------------- ( -22.50) >DroPse_CAF1 94581 86 + 1 UUCAAUAAGCGCAUUGGGGCAUGUGAUUACGCUGCCGC---CUCACAUGUGACGAGGCUCUCAUGCCGCGUCAUGGCA-CGCCA-----AGCGCC------------ ........((((.((((((((.(((....)))))))((---(......((((((.(((......))).))))))))).-..)))-----))))).------------ ( -37.60) >DroGri_CAF1 122104 85 + 1 UUCAAUAAAUUCAGUGGGGCAUGUGAUUACGCAAAAACA----CACAUGUGGCGGUGAUCACAUGCCACGUCAUGCUGCCACCG---ACUCC--------------- ..........((.(((((((((((((((((((....(((----....))).)).)))))))))))))..(.....)..)))).)---)....--------------- ( -29.80) >DroSec_CAF1 86039 96 + 1 UUCAAUAAAUUCACUGGCCCAUGUGAUUAUGCUGGGGCG--CUCACAUGUGCCUCCGCUCACAUGCCUCGUCAUGCCA-CGCCC-CCCAUCCGCCAGUC-------G ............((((((.(((((((....((.((((((--(......))))))).)))))))))...(((......)-))...-.......)))))).-------. ( -31.70) >DroEre_CAF1 89103 102 + 1 UUCAAUAAAUUCACUGGGCCAUGUGAUUACGCCGUGGCG--CUCACAUGUGCCUUCGCUCACAUGCCGCGUCAUGCCA-CGCCC--CCAUCCGCCAGUCUCCACCCG ............(((((((.((((((....((...((((--(......)))))...)))))))))))((((......)-)))..--........))))......... ( -28.90) >consensus UUCAAUAAAUUCACUGGGCCAUGUGAUUACGCUGAAGCG__CUCACAUGUGCCGCCGCUCACAUGCCACGUCAUGCCA_CGCCA___CAUCCGCC____________ ...............((..(((((((....((....))....)))))))............((((......))))......))........................ ( -9.23 = -9.90 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:37 2006