
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,264,473 – 4,264,596 |
| Length | 123 |
| Max. P | 0.727084 |

| Location | 4,264,473 – 4,264,564 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.66 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -17.52 |
| Energy contribution | -19.28 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4264473 91 + 27905053 GUAGGGGCAG------------------GAGGCC-ACGGGCCGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCUCUCUCGGACCGGAUUUUGUUUGCCUGACUUUGU ((..((((((------------------(.((((-...))))(((((((((.(((....)))(((((.........)))))....))))))))).))))))).))..... ( -33.10) >DroSec_CAF1 66172 92 + 1 GUAGGGGCAG------------------GAGGCCCCCGGGCAGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCUCUCUCGGACCGGAUUUUGUUUGCCUGACUUUGU ...(((((..------------------...)))))(((((((((((((((.(((....)))(((((.........)))))....))))))))...)))))))....... ( -37.10) >DroSim_CAF1 66035 90 + 1 GUAGGGGUAG------------------GAGGCCCCCGGGCAGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCU--CUCGGACCGGAUUUUGUUUGCCUGACUUUGU ...(((((..------------------...)))))(((((((((((((((.(((....))).((((.........)--)))...))))))))...)))))))....... ( -35.30) >DroEre_CAF1 69074 106 + 1 GGACGAG-AGCUGCAGCUGGAGAACCUGAAGG-CCCCGGGCAGGAGUCCGGAGUCAAUAGAGAGAGAUAUAUCUCCU--CUCGGACCGGAUUUUGUUUGCCUGACUUUGU ..(((((-.(((....(.((....)).)..))-)..(((((((((((((((..((...((((.((((....))))))--))..))))))))))...)))))))..))))) ( -34.30) >DroAna_CAF1 68825 78 + 1 CUAGGCGAAA------------------GAAGUCUU-------UAGCUUGCAGUCCAUAGACAGAGAUAUAUCUC-------UGGUCGGAUUUUGUUUGGCUGACUUUGU ..........------------------((((((..-------.((((.(((((((.....((((((....))))-------))...))))..)))..)))))))))).. ( -22.10) >consensus GUAGGGGCAG__________________GAGGCCCCCGGGCAGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCU__CUCGGACCGGAUUUUGUUUGCCUGACUUUGU ............................((((....(((((((((((((((.(((....))).((((....))))..........)))))))))...)))))).)))).. (-17.52 = -19.28 + 1.76)

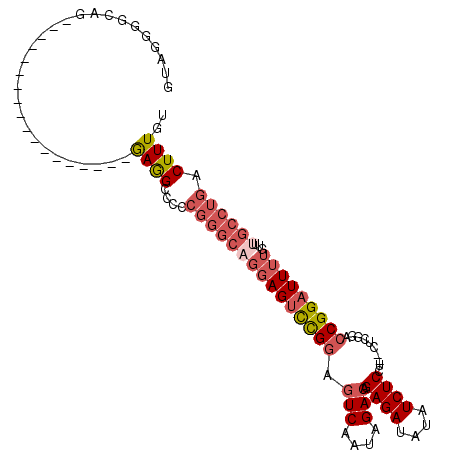

| Location | 4,264,486 – 4,264,596 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4264486 110 + 27905053 GCC-ACGGGCCGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCUCUCUCGGACCGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCCGGA .((-..((((((((((((((.(((....)))(((((.........)))))....))))))))))...))))(((...(((((((.(((....))))))))))..))).)). ( -40.50) >DroPse_CAF1 74101 84 + 1 ---------CCGGA----GGAGUCUAUAGAUA--CAUAUAUCUGCU---------CGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCU--- ---------..((.----(((((((.((((((--....))))))..---------.))))))).....)).(((...(((((((.(((....))))))))))..))).--- ( -22.90) >DroSec_CAF1 66185 111 + 1 GCCCCCGGGCAGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCUCUCUCGGACCGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCUGGA ....((((((((((((((((.(((....)))(((((.........)))))....)))))))....(((((.....((((((.(....).)))))).)))))))))))))). ( -42.90) >DroEre_CAF1 69104 108 + 1 G-CCCCGGGCAGGAGUCCGGAGUCAAUAGAGAGAGAUAUAUCUCCU--CUCGGACCGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCCGGA .-..((((((((((((((((..((...((((.((((....))))))--))..)))))))))....(((((.....((((((.(....).)))))).)))))))))))))). ( -47.80) >DroYak_CAF1 68034 108 + 1 G-CCCCGAGCAGGAGUCCGAAGUCAAUAGACAGAGAUAUAUCUUCU--CUCGGACCGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCCGGA .-..(((..((((.((((((.(((....))).((((.......)))--)))))))..........(((((.....((((((.(....).)))))).)))))))))..))). ( -39.60) >DroPer_CAF1 74563 84 + 1 ---------CCGGA----GGAGUCUAUAGAUA--CAUAUAUCUGCU---------CGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCU--- ---------..((.----(((((((.((((((--....))))))..---------.))))))).....)).(((...(((((((.(((....))))))))))..))).--- ( -22.90) >consensus G_C_CCGGGCAGGAGUCCGGAGUCAAUAGACAGAGAUAUAUCUCCU__CUCGGACCGGAUUUUGUUUGCCUGACUUUGUUGCCACUUGCCAACAAUGGCAACCUGUCCGGA ..................((((...(((........)))..)))).........((((((...(.(((((.....((((((.(....).)))))).))))).).)))))). (-20.50 = -20.83 + 0.33)



| Location | 4,264,486 – 4,264,596 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4264486 110 - 27905053 UCCGGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCGGUCCGAGAGAGGAGAUAUAUCUCUGUCUAUUGACUCCGGACUCCGGCCCGU-GGC .((.(((..(((((((((.......)))))))))...))).))....((....((((((((.(((.(((((....))))).((....)))))))))..))))....)-).. ( -38.30) >DroPse_CAF1 74101 84 - 1 ---AGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCG---------AGCAGAUAUAUG--UAUCUAUAGACUCC----UCCGG--------- ---.(((..(((((((((.......)))))))))...))).............(((---------((.((.((((..--....))))..)).)----).)))--------- ( -21.10) >DroSec_CAF1 66185 111 - 1 UCCAGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCGGUCCGAGAGAGGAGAUAUAUCUCUGUCUAUUGACUCCGGACUCCUGCCCGGGGGC (((.(((..(((((((((.......)))))))))...))).((((.......(((((..(((.((((((((....))))).))).)))...)))))....)))).)))... ( -39.40) >DroEre_CAF1 69104 108 - 1 UCCGGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCGGUCCGAG--AGGAGAUAUAUCUCUCUCUAUUGACUCCGGACUCCUGCCCGGGG-C (((((.(((((((((.((((((.(....).)))))).....)))))......(((((((..((--((((((....)))).))))...))..)))))..)))).))))).-. ( -42.60) >DroYak_CAF1 68034 108 - 1 UCCGGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCGGUCCGAG--AGAAGAUAUAUCUCUGUCUAUUGACUUCGGACUCCUGCUCGGGG-C ((((..(((((((((.((((((.(....).)))))).....))))).........((((((((--(((.......)))).(((....))).)))))))))))..)))).-. ( -40.10) >DroPer_CAF1 74563 84 - 1 ---AGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCG---------AGCAGAUAUAUG--UAUCUAUAGACUCC----UCCGG--------- ---.(((..(((((((((.......)))))))))...))).............(((---------((.((.((((..--....))))..)).)----).)))--------- ( -21.10) >consensus UCCAGACAGGUUGCCAUUGUUGGCAAGUGGCAACAAAGUCAGGCAAACAAAAUCCGGUCCGAG__AGGAGAUAUAUCUCUGUCUAUUGACUCCGGACUCCGGCCCGG_G_C ....(((..(((((((((.......)))))))))...)))...............((((((.......(((((......)))))........))))))............. (-21.61 = -22.28 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:32 2006