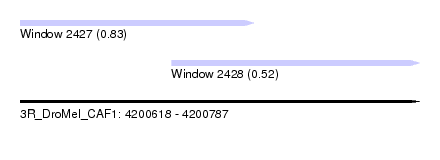
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,200,618 – 4,200,787 |
| Length | 169 |
| Max. P | 0.827454 |

| Location | 4,200,618 – 4,200,717 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.93 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
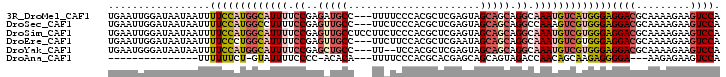
>3R_DroMel_CAF1 4200618 99 + 27905053 UGAAUUGGAUAAUAAUUUUCCAUGGCAUUUUCCGAGAUGCC---UUUUCCCACGCUCGAGUAGCAGCAGGCAAAUGUCAUGGGAGGACGCAAAAGAAGUCCA .................(((((((((((((.((((((....---.))))....((((.....).))).)).)))))))))))))((((.........)))). ( -29.60) >DroSec_CAF1 11985 99 + 1 UGAAUUGGAUAAUAAUUUUCCAUGGCCUUUUCCGAGUUGCC---UUCUCCCACGCUCGAGUAGCAGCAGGCCAAAGUCGUGGGAGGACGCAAAAGAAGUCCA .....((((.........))))(((.(((((.....((((.---(((((((((((((.....).))).(((....)))))))))))).)))).))))).))) ( -33.10) >DroSim_CAF1 12168 102 + 1 UGAAUUGGAUAAUAAUUUUCCAUGGCAUUUUCCGAGUUGCCUCCUUCUCCCACGCUCGAGUAGCAGCAGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCA .................(((((((((((((.((..(((((......(((........)))..))))).)).)))))))))))))((((.........)))). ( -32.10) >DroEre_CAF1 12693 99 + 1 UGAAUUGGAUAAUAAUUUUCCCUGGCAUUUUCCGAGUUGCC---UUCUUCCACGCUCGAAUAGCAGCAGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCA .((((((.....))))))((((((((((((.((..(((((.---(((..........)))..))))).)).)))))))).))))((((.........)))). ( -28.80) >DroYak_CAF1 12360 97 + 1 UGAAUGGGAUAAUAAUUUUCCAUGGCAUUUUCCGAGCUGCC---UU--UCCACGCUCGAGUAGCAGCAGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCA .................(((((((((((((.((..(((((.---.(--((.......)))..))))).)).)))))))))))))((((.........)))). ( -35.50) >DroAna_CAF1 14051 79 + 1 ---------------UUUUUUCU-GUAUUUUCCCC-ACACA---UUUUCCCACGCACGAGCAGCAGUAGACCAACAGCAAGAGGGGA---AAGAGAAGUCCA ---------------...(((((-...((((((((-.....---.........((....)).((.((......)).))....)))))---)))))))).... ( -16.90) >consensus UGAAUUGGAUAAUAAUUUUCCAUGGCAUUUUCCGAGUUGCC___UUCUCCCACGCUCGAGUAGCAGCAGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCA .................(((((((((((((.((..(((((......................))))).)).)))))))))))))((((.........)))). (-20.57 = -21.93 + 1.37)
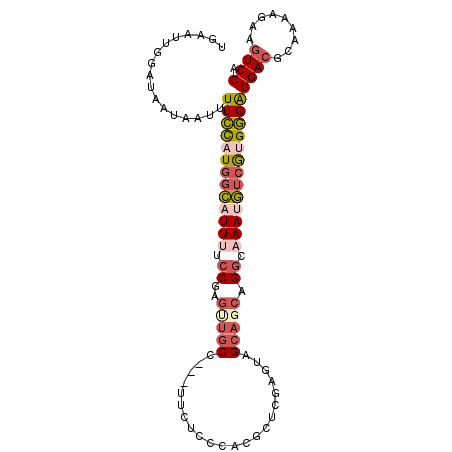
| Location | 4,200,682 – 4,200,787 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -30.99 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4200682 105 + 27905053 AGGCAAAUGUCAUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCCGGGGCAUUAUGUGUCCUUUCCCGGACUCGAGAC-CGUCCGCCUGU ((((.........(((((((((((......(((........)))..........((((((....)))))))))))))..))))((((.......-.)))))))).. ( -37.70) >DroSec_CAF1 12049 105 + 1 AGGCCAAAGUCGUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCCGGAGCAUUAUGUGUUCUUUCCCGGACUCGAGAC-CGUCCGCCUGU ((((.........(((((((((((......(((........)))..........((((((....)))))))))))))..))))((((.......-.)))))))).. ( -32.00) >DroSim_CAF1 12235 105 + 1 AGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCCGGAGCAUUAUGUGUUCUUUCCCGGACUCGAGAC-CGUCCGCCUGU ((((.........(((((((((((......(((........)))..........((((((....)))))))))))))..))))((((.......-.)))))))).. ( -32.90) >DroEre_CAF1 12757 106 + 1 AGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCUGGGGCAUUAUGUGUUCUUUCCCUGACUCGAGACUCGUUCGCCUGU ((((....((((.(((((((((((.((....(((((((...................)))))))...)).)))))))..))))))))..(((.....))))))).. ( -32.91) >DroYak_CAF1 12422 105 + 1 AGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCUGGGGCAUUAUGUGUUCUUUCCCGGACUCGAGAC-CGUUCGUCUGU ((((....(((.((((((((((((.((....(((((((...................)))))))...)).)))))))..))))))))..(((..-..))))))).. ( -30.21) >DroAna_CAF1 14098 97 + 1 AGACCAACAGCAAGAGGGGA---AAGAGAAGUCCACCCAGAGAUGUAAUAAAAUUAAUGCCAAGGCAUUAUAUGUUCUUUUCCAAACU-GA-----GUCGUCCUGC .(((...(((......((((---(((((..(((........)))..........((((((....))))))....)))))))))...))-).-----)))....... ( -20.20) >consensus AGGCAAAUGUCGUGGGAGGACGCAAAAGAAGUCCAGCAGAAGACAUAAUAAAAUUAAUGCCGGGGCAUUAUGUGUUCUUUCCCGGACUCGAGAC_CGUCCGCCUGU ((((.........(((((((((((......(((........)))..........((((((....)))))))))))))))))..((((.........)))))))).. (-24.67 = -25.03 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:07 2006