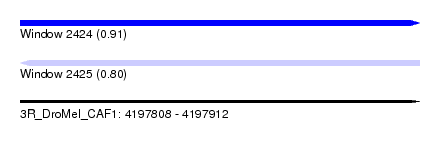
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,197,808 – 4,197,912 |
| Length | 104 |
| Max. P | 0.914109 |

| Location | 4,197,808 – 4,197,912 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -19.96 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4197808 104 + 27905053 AGUCGACAGUC-GGC-GGUUUGGCCUGCGUUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAACGCAGAAUUGAUGGCCAAAAGGGG---CUGUGCCA------- .((((.....)-)))-....(((((((((((((.......((((...(((((((....))))))))))).))))))))).....((((((......))---))))))))------- ( -39.50) >DroVir_CAF1 25026 103 + 1 AGAGGACACUCCGGC-GGC---GACGGCGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAAAGCGGAAUUGAUGGUCAGACAAGGU--GAGGGUCA------- ....(((....((.(-(..---..)).))((..((((((((((...((((((((....)))))))).((((((........))))))).))))))))).--.)).))).------- ( -34.70) >DroGri_CAF1 13624 99 + 1 AGACGA-----CGGCAAAC---GAUGGCGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAAAGAAGAAUUGAUGGUCAAACAAGGU--GUGUGGCA------- ......-----((((((((---((.(((....))))))))))))).((((((((....)))))))).((((((........))))))((((.((.....--)).)))).------- ( -29.90) >DroMoj_CAF1 12892 103 + 1 AGCGGGCACUCCGGC-GGC---GACGGUGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAACGAAGAAUUGAUGGCCGAACAAGGU--GCGGGGCU------- (((..(((((....(-(((---.(((((.....(((((((((.((.((((((((....)))))))).)).))))))))).)))).).)))).....)))--))...)))------- ( -36.70) >DroAna_CAF1 11008 110 + 1 AGUCGACAGUC-GGC-GGU-UGGCCGGCGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAACGCAGAAUUGAUGGCCAAACGAGA---CCGUACGACUGGCAG (((((((.(((-..(-(.(-((((((...(..((((((((((.((.((((((((....)))))))).)).)))..))))))).).))))))).)).))---).)).)))))..... ( -42.20) >DroPer_CAF1 10260 95 + 1 AGUCGACGGCC-G-------------CUGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAACGCUGAAUUGAUGGCCAAACGAGACGACCGUGCGA------- ..(((((((.(-(-------------...(((((((.(((((.((.((((((((....)))))))).)).)))))(((........))).))))))).)).)))).)))------- ( -32.80) >consensus AGUCGACAGUC_GGC_GGC___GACGGCGCUUGUUUUGUUUGCUGAUUGCGCAAAUAUUUGCGCAGGCAUUAAACGCAGAAUUGAUGGCCAAACAAGGU__CCGUGCCA_______ .............................(((((((...........(((((((....)))))))((((((((........))))).))))))))))................... (-19.96 = -19.10 + -0.86)

| Location | 4,197,808 – 4,197,912 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4197808 104 - 27905053 -------UGGCACAG---CCCCUUUUGGCCAUCAAUUCUGCGUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAACGCAGGCCAAACC-GCC-GACUGUCGACU -------((((...(---((......)))........((((((((..(((.(((((((....)))))))....)))........))))))))))))....-..(-(.....))... ( -28.60) >DroVir_CAF1 25026 103 - 1 -------UGACCCUC--ACCUUGUCUGACCAUCAAUUCCGCUUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCGCCGUC---GCC-GCCGGAGUGUCCUCU -------.(((.(((--...(((.((((...........((.......)).(((((((....))))))).))))))).........(((....)---)).-....))).))).... ( -23.50) >DroGri_CAF1 13624 99 - 1 -------UGCCACAC--ACCUUGUUUGACCAUCAAUUCUUCUUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCGCCAUC---GUUUGCCG-----UCGUCU -------.((.((..--...(((((((..(((.((........)).)))..(((((((....))))))).....)))))))..((((((....)---)))))..)-----).)).. ( -19.30) >DroMoj_CAF1 12892 103 - 1 -------AGCCCCGC--ACCUUGUUCGGCCAUCAAUUCUUCGUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCACCGUC---GCC-GCCGGAGUGCCCGCU -------(((...((--((....((((((............((((..((..(((((((....)))))))..))..)))).......((......---)).-))))))))))..))) ( -29.00) >DroAna_CAF1 11008 110 - 1 CUGCCAGUCGUACGG---UCUCGUUUGGCCAUCAAUUCUGCGUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCGCCGGCCA-ACC-GCC-GACUGUCGACU .....(((((.((((---((.((.((((((..((....)).((((..((..(((((((....)))))))..))..))))............)))))-).)-)..-))))))))))) ( -38.60) >DroPer_CAF1 10260 95 - 1 -------UCGCACGGUCGUCUCGUUUGGCCAUCAAUUCAGCGUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCAG-------------C-GGCCGUCGACU -------(((.(((((((.((.(((((((..........))((((..((..(((((((....)))))))..))..))))....)))))))-------------)-))))))))).. ( -34.40) >consensus _______UGGCACGC__ACCUCGUUUGGCCAUCAAUUCUGCGUUUAAUGCCUGCGCAAAUAUUUGCGCAAUCAGCAAACAAAACAAGCGCCGUC___GCC_GCC_GACUGUCGACU .........................................((((..((..(((((((....)))))))..))..))))..................................... (-11.90 = -12.23 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:04 2006