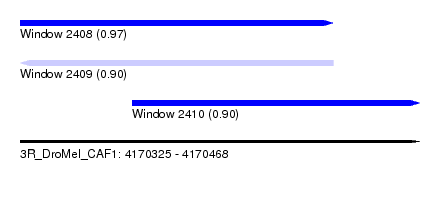
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,170,325 – 4,170,468 |
| Length | 143 |
| Max. P | 0.968446 |

| Location | 4,170,325 – 4,170,437 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.01 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -14.93 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4170325 112 + 27905053 UGGGCUCUGCCAGUUCUUCACGUUUUCGGGCUCCAUUUUAGCCAUAGUUUUGUUUUCUA--UA---UUCUUGUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUU ..(((...))).........((....))((((.......))))........((((((((--((---....))))).)))))((((((.((((((.....---..)))))).)).)))).. ( -29.70) >DroSim_CAF1 20726 112 + 1 UGGGCUCUGCCAGUUCUUCACGUUUUCGGGCUCCAUAUUAGCCAUAGUUUUGUUUUCUA--UA---UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUU ..(((...))).........((....))((((.......))))........((((((((--..---......))).)))))((((((.((((((.....---..)))))).)).)))).. ( -27.30) >DroEre_CAF1 10110 106 + 1 ----AUCUGACGGUUCUUCACGUUUUCGGUCUCCAUUUUAGCCAUAGUUUUGUUUUCUA--UA---UUCUUAAG--ACAAUGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUU ----....((((........))))...............((((...((.(((((((...--..---.....)))--)))).)).(((.((((((.....---..)))))).))).)))). ( -22.20) >DroWil_CAF1 6628 100 + 1 ---UAAUUACCCCUACUCCCCAUUUUUGGGG----------AGAAAUGUUUGUUUU-UAAGUUUUCUUCUUUUU------CUCCACAUUGCUGCCAUAUUGUCUUCAGCUCUGUUGGAAC ---............(((((((....)))))----------)).............-.................------.((((((..((((............))))..)).)))).. ( -22.80) >DroYak_CAF1 20411 112 + 1 UGGUAUAUGACGGUCCUUUACGUUUUCGGGCUCCAUCUCAGCCAUAGUUUUGUUUUCUA--UA---UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGUUU ((((..(((..(((((...........))))).)))....)))).......((((((((--..---......))).)))))((((((.((((((.....---..)))))).)).)))).. ( -25.40) >consensus UGGGAUCUGCCAGUUCUUCACGUUUUCGGGCUCCAUUUUAGCCAUAGUUUUGUUUUCUA__UA___UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU___CUGCAGUUUAGCCGGCUU ....................((....))((((.......))))......................................((((((.((((((..........)))))).)).)))).. (-14.93 = -14.46 + -0.47)



| Location | 4,170,325 – 4,170,437 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.01 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -14.36 |
| Energy contribution | -13.76 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4170325 112 - 27905053 AAGCCGGCUAAACUGCAG---AUGUGGCAGUUAAGCGGCGUUUUACCACAAGAA---UA--UAGAAAACAAAACUAUGGCUAAAAUGGAGCCCGAAAACGUGAAGAACUGGCAGAGCCCA ..((((.((.((((((..---.....)))))).))))))(((((..(((.....---.(--(((.........))))((((.......)))).......))).))))).(((...))).. ( -28.10) >DroSim_CAF1 20726 112 - 1 AAGCCGGCUAAACUGCAG---AUGUGGCAGUUAAGCGGCGUUUUACCAAAAGAA---UA--UAGAAAACAAAACUAUGGCUAAUAUGGAGCCCGAAAACGUGAAGAACUGGCAGAGCCCA ..(((((((.((((((..---.....)))))).))).(((((((..........---.(--(((.........))))((((.......))))..))))))).......))))........ ( -27.80) >DroEre_CAF1 10110 106 - 1 AAGCCGGCUAAACUGCAG---AUGUGGCAGUUAAGCGGCAUUGU--CUUAAGAA---UA--UAGAAAACAAAACUAUGGCUAAAAUGGAGACCGAAAACGUGAAGAACCGUCAGAU---- ..((((.((.((((((..---.....)))))).))))))...((--(((.....---((--(((.........))))).........)))))........................---- ( -22.14) >DroWil_CAF1 6628 100 - 1 GUUCCAACAGAGCUGAAGACAAUAUGGCAGCAAUGUGGAG------AAAAAGAAGAAAACUUA-AAAACAAACAUUUCU----------CCCCAAAAAUGGGGAGUAGGGGUAAUUA--- .(((((.((..((((....(.....).))))..)))))))------.................-.......((....((----------(((((....))))))).....)).....--- ( -24.10) >DroYak_CAF1 20411 112 - 1 AAACCGGCUAAACUGCAG---AUGUGGCAGUUAAGCGGCGUUUUACCAAAAGAA---UA--UAGAAAACAAAACUAUGGCUGAGAUGGAGCCCGAAAACGUAAAGGACCGUCAUAUACCA ...((.(((.((((((..---.....)))))).))).(((((((..........---.(--(((.........))))((((.......))))..)))))))...)).............. ( -24.30) >consensus AAGCCGGCUAAACUGCAG___AUGUGGCAGUUAAGCGGCGUUUUACCAAAAGAA___UA__UAGAAAACAAAACUAUGGCUAAAAUGGAGCCCGAAAACGUGAAGAACCGGCAGAUACCA ..((((.((.((((((..........)))))).))))))................................................................................. (-14.36 = -13.76 + -0.60)



| Location | 4,170,365 – 4,170,468 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.39 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -13.09 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4170365 103 + 27905053 GCCAUAGUUUUGUUUUCUA--UA---UUCUUGUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUUGC--UGGAGAUAAAAUUGCAUU---AUCAAGU----UUUC .(((.......((((((((--((---....))))).)))))((((((.((((((.....---..)))))).)).))))....--))).(((((.......))---)))....----.... ( -26.60) >DroSim_CAF1 20766 106 + 1 GCCAUAGUUUUGUUUUCUA--UA---UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUUGC--UGGAGAUAAAAUUGCAUCAUCAUCAAGU----UUUC ((...((((((((((((..--..---.......(((((...((((((.((((((.....---..)))))).)).))))))))--))))))))))))))).............----.... ( -25.10) >DroEre_CAF1 10146 104 + 1 GCCAUAGUUUUGUUUUCUA--UA---UUCUUAAG--ACAAUGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGCUUGC--UAGAGAUAAAAUUGCAUUAUCAUCAAGU----UUUC ((...((((((((((.(((--..---.((....)--)(((.((((((.((((((.....---..)))))).)).))))))).--))))))))))))))).............----.... ( -23.50) >DroWil_CAF1 6656 110 + 1 -AGAAAUGUUUGUUUU-UAAGUUUUCUUCUUUUU------CUCCACAUUGCUGCCAUAUUGUCUUCAGCUCUGUUGGAACAC--CUAAGAAAAAUAUGCAUAAUCAUCAUAUGUAUGUUC -((((((....)))))-)...(((((((......------.((((((..((((............))))..)).))))....--..)))))))(((((((((.......))))))))).. ( -18.92) >DroYak_CAF1 20451 106 + 1 GCCAUAGUUUUGUUUUCUA--UA---UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU---CUGCAGUUUAGCCGGUUUGC--UAGAGAUAAAGUUGCAUUAUCAUCAGUU----UUCC ......((..(((...((.--((---((..((((((((...((((((.((((((.....---..)))))).)).))))))))--)))))))).))..)))..))........----.... ( -22.70) >DroAna_CAF1 17242 102 + 1 -----AGUCUUGCACUCCAAUUA---UUCUUUUGGUUAUAACCCGCAUAUCUGCCACAU---CG---GAUUUGUCGGAUCACCCUGAGAAAGAAAGUGCAUCUUCAUCAAGU----UUUC -----.....((((((....((.---(((((..(((.((...(((((.(((((......---))---))).)).))))).)))..))))).)).))))))............----.... ( -21.80) >consensus GCCAUAGUUUUGUUUUCUA__UA___UUCUUUUGGUAAAACGCCGCUUAACUGCCACAU___CUGCAGUUUAGCCGGCUUGC__UGGAGAUAAAAUUGCAUUAUCAUCAAGU____UUUC .....(((((((((((............(....).......((((((.((((((..........)))))).)).))))........)))))))))))....................... (-13.09 = -12.48 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:49 2006