
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,163,524 – 4,163,657 |
| Length | 133 |
| Max. P | 0.654786 |

| Location | 4,163,524 – 4,163,620 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -25.69 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4163524 96 + 27905053 CGGCGGAGC------------GGGGGCAUUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA .........------------.(((((.((.((((.(....)((((..((((((((((.((((.((....))...))))))))).))))))))))))).)).))))). ( -33.80) >DroSec_CAF1 7485 108 + 1 CGUCGGAGAGGGAGAGGGGGGGGGGGCACUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA ....((((.(...(((..(.((.(((((.((((((.(((..(((...(((..(((....)))...)))..)))..)))))))))))))).)).).)))...).)))). ( -35.00) >DroSim_CAF1 13697 104 + 1 CGUCGGAGAGGGGAAGG----GGGGGCAUUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA (.((.....)).)....----.(((((.((.((((.(....)((((..((((((((((.((((.((....))...))))))))).))))))))))))).)).))))). ( -34.00) >DroEre_CAF1 1000 94 + 1 AGGGGAA--------------GGGGACGUUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA .......--------------((((.((((.((((.(....)((((..((((((((((.((((.((....))...))))))))).))))))))))))).)))))))). ( -35.90) >DroYak_CAF1 13541 94 + 1 CGGAGGA--------------GGGGGCGUUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA .......--------------.((((((((.((((.(....)((((..((((((((((.((((.((....))...))))))))).))))))))))))).)))))))). ( -39.50) >DroAna_CAF1 7810 76 + 1 ------A--------------GGAGACGAUGAAGCUGGGGACG------------GAGACGAAAGCGGCAGCGAGUUUGCUUCAUGCUCGUUGCACUUGAACGCUUCA ------.--------------(....)..((((((......((------------....)).(((..(((((((((.((...)).))))))))).)))....)))))) ( -27.70) >consensus CGGCGGA______________GGGGGCAUUGAAGUUGGGGACGCAAAUCGAGUUGGAGACGAAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCA .............................((((((.(....).........(((.(((.(....)..(((((((((.........))))))))).))).))))))))) (-25.69 = -26.05 + 0.36)


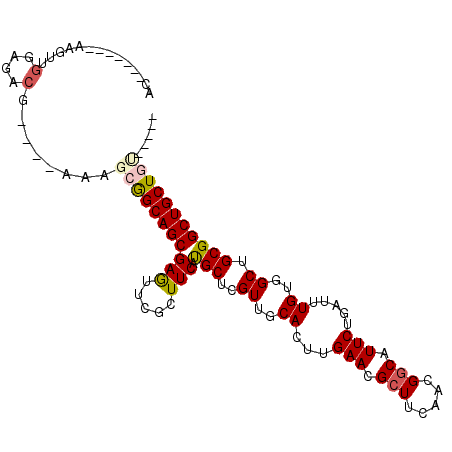
| Location | 4,163,552 – 4,163,657 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4163552 105 + 27905053 ACGCAAAUCGAGUUGGAGACG----AAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUGUGCUGU .(((((((((.(((((((.((----.(((..(((((((((.........))))))))).)))...))))))))).......)))))))))...(((((......))))) ( -38.90) >DroVir_CAF1 4545 100 + 1 ------ACGUAGCCGGAGACUGGCAGCCGCAGCAGCGAAUUCGCUUCACGCCGGUUGCACUUGAAUGCUUCAACGACAUUCUGAUUUGUGGCUGCGGCUGCUGCCA--- ------.....((.(....).((((((((((((((((....)))).(((((.....))....((((((......).)))))......)))))))))))))))))..--- ( -40.20) >DroPse_CAF1 14586 92 + 1 GG-------AAGUUGGAGACG----ACGAAGGCAGCGAGUUCUCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUA------ .(-------..(((((((.((----...((((((((((((.........))))))))).)))...)))))))))..)..........(..((....))..)..------ ( -31.70) >DroSim_CAF1 13733 105 + 1 ACGCAAAUCGAGUUGGAGACG----AAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUGUGCUGC .(((((((((.(((((((.((----.(((..(((((((((.........))))))))).)))...))))))))).......)))))))))...(((((......))))) ( -40.10) >DroAna_CAF1 7830 89 + 1 ACG------------GAGACG----AAAGCGGCAGCGAGUUUGCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUGU----U .((------------....))----..(((((((((((((.((...)).))))((..((...(((.(((.....))).))).....))..))....))))))))----) ( -30.90) >DroPer_CAF1 14481 92 + 1 GG-------AAGUUGGAGACG----ACGAAGGCAGCGAGUUCUCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUA------ .(-------..(((((((.((----...((((((((((((.........))))))))).)))...)))))))))..)..........(..((....))..)..------ ( -31.70) >consensus AC_______AAGUUGGAGACG____AAAGCGGCAGCGAGUUCGCUUCAUGCUCGUUGCACUUGAACGCUUCAACGGCAUUCUGAUUUGUGGCUGCGGCUGCUGU_____ ..............(....)........(((((((((((.....))).(((..((..((...(((.(((.....))).))).....))..)).)))))))))))..... (-22.83 = -23.33 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:43 2006