
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,148,659 – 4,148,774 |
| Length | 115 |
| Max. P | 0.992262 |

| Location | 4,148,659 – 4,148,755 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.24 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
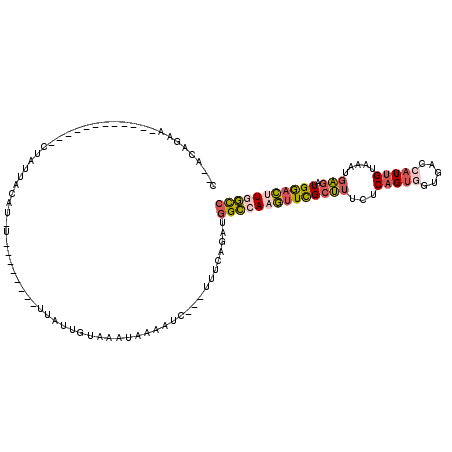
>3R_DroMel_CAF1 4148659 96 + 27905053 CAUACAAAA------------CCAUUACAU-U--------UUAUUGUAAACAAAAUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCC .........------------...(((((.-.--------....))))).....(((---.....)))((((((((((((((...((((((...)))))).....))).))))))))))) ( -27.70) >DroSec_CAF1 9618 96 + 1 CUUACAGAA------------CUAUUACAU-U--------UUAUUGUUAACAAACUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCC .........------------.........-.--------.................---........((((((((((((((...((((((...)))))).....))).))))))))))) ( -24.80) >DroEre_CAF1 9782 104 + 1 GCCACAGAAUAAAUGUAAAAAGUAUU-----A--------CUAUUGUUAAUGCCAUC---UUUUAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCC (((....(((((..((((......))-----)--------)..)))))...((((((---.....)))))).((((((((((...((((((...)))))).....))).)))))))))). ( -28.50) >DroWil_CAF1 18553 99 + 1 C---CAGAA------------GUUUUCCAUUCAAGUCCCUUAAUUGUC----CUUUC--UUUGCAGAUGGCCAAACUUGCUUUUUCAGUCGUCACAAUUGUCGAUCAGAUGAAUCUAGCC .---...((------------((((.(((((((((.............----.....--))))..)))))..))))))(((..((((((((.((....)).))))....))))...))). ( -14.27) >DroYak_CAF1 10072 108 + 1 UCCACAGAAUAUUUAUAAAAAAUAAUCAAU-U--------UUCUUGUCAAUGAAAUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCC ......((.(((((.....))))).))...-.--------.....(((..((((...---.)))))))((((((((((((((...((((((...)))))).....))).))))))))))) ( -27.50) >DroAna_CAF1 11088 100 + 1 -----UUAACACUUUU-------AGUUCAUUU--------GUGUUCUACAUUAUUUUAUUCCACAAAUGGUCAAGUUUGCCUUUUCAGUCUUUACACUCGUUCAGGGAAUGGACCUAAUG -----..(((((....-------.........--------)))))...(((((.((((((((...((((((.(((.(((......))).)))...)).))))...))))))))..))))) ( -14.52) >consensus C__ACAGAA____________CUAUUACAU_U________UUAUUGUAAAUAAAAUC___UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCC ....................................................................((((((((((((((...(((((.....))))).....))).))))))))))) (-15.36 = -16.03 + 0.67)

| Location | 4,148,678 – 4,148,774 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.00 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4148678 96 + 27905053 UUAUUGUAAACAAAAUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCCGAGAAAUUG----AGAAG----GGAGU------------- ...............((---((((....((((((((((((((...((((((...)))))).....))).)))))))))))((....)).----.))))----))...------------- ( -31.20) >DroSec_CAF1 9637 96 + 1 UUAUUGUUAACAAACUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCCGAGAAAUUG----AGAAG----GGAGU------------- .............((((---((((....((((((((((((((...((((((...)))))).....))).)))))))))))((....)).----.))).----)))))------------- ( -31.80) >DroEre_CAF1 9809 96 + 1 CUAUUGUUAAUGCCAUC---UUUUAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCCGAGAAAUUG----AGAAG----GGAGU------------- ..........(.((.((---((.....(((((((((((((((...((((((...)))))).....))).)))))))))))).......)----))).)----).)..------------- ( -31.40) >DroWil_CAF1 18578 110 + 1 UAAUUGUC----CUUUC--UUUGCAGAUGGCCAAACUUGCUUUUUCAGUCGUCACAAUUGUCGAUCAGAUGAAUCUAGCCGACAAAAUGUCUUAAAAUGAUUUUAGCGAGAAUC----AA ...(((.(----(..((--(....))).)).))).((((((...(((......(((.((((((...(((....)))...))))))..))).......)))....))))))....----.. ( -20.52) >DroYak_CAF1 10103 96 + 1 UUCUUGUCAAUGAAAUC---UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCCGAGAAAUUG----AGAAG----GGAGU------------- (((((.(((((...(((---.....)))((((((((((((((...((((((...)))))).....))).))))))))))).....))))----).)))----))...------------- ( -34.70) >DroAna_CAF1 11108 112 + 1 GUGUUCUACAUUAUUUUAUUCCACAAAUGGUCAAGUUUGCCUUUUCAGUCUUUACACUCGUUCAGGGAAUGGACCUAAUGGAAAAAUUU----AAAAA----CUAGAGAGCUCCCGAAAA ..(((((...(((..((.(((((.....((((....((.(((...((((......))).)...))).))..))))...))))).))..)----))...----....)))))......... ( -18.80) >consensus UUAUUGUAAAUAAAAUC___UUUCAGAUGGCCAAGUUCGCUUUCUCAGUGGUGACCAUUGUAAAUGAGAUGGACUUGGCCGAGAAAUUG____AGAAG____GGAGU_____________ ...........................(((((((((((((((...(((((.....))))).....))).))))))))))))....................................... (-17.21 = -17.88 + 0.67)


| Location | 4,148,678 – 4,148,774 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.00 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -11.76 |
| Energy contribution | -11.32 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4148678 96 - 27905053 -------------ACUCC----CUUCU----CAAUUUCUCGGCCAAGUCCAUCUCAUUUACAAUGGUCACCACUGAGAAAGCGAACUUGGCCAUCUGAAA---GAUUUUGUUUACAAUAA -------------.....----.....----...((((..((((((((...((.(.(((.((.(((...))).)).))).).))))))))))....))))---................. ( -18.10) >DroSec_CAF1 9637 96 - 1 -------------ACUCC----CUUCU----CAAUUUCUCGGCCAAGUCCAUCUCAUUUACAAUGGUCACCACUGAGAAAGCGAACUUGGCCAUCUGAAA---GAGUUUGUUAACAAUAA -------------.....----...((----(..((((..((((((((...((.(.(((.((.(((...))).)).))).).))))))))))....))))---))).............. ( -19.40) >DroEre_CAF1 9809 96 - 1 -------------ACUCC----CUUCU----CAAUUUCUCGGCCAAGUCCAUCUCAUUUACAAUGGUCACCACUGAGAAAGCGAACUUGGCCAUCUAAAA---GAUGGCAUUAACAAUAG -------------.....----....(----(..((((((((....(.((((..........)))).)....))))))))..)).....((((((.....---))))))........... ( -22.60) >DroWil_CAF1 18578 110 - 1 UU----GAUUCUCGCUAAAAUCAUUUUAAGACAUUUUGUCGGCUAGAUUCAUCUGAUCGACAAUUGUGACGACUGAAAAAGCAAGUUUGGCCAUCUGCAAA--GAAAG----GACAAUUA .(----((((.((((((((............(((.((((((..((((....))))..))))))..)))..(.((.....)))...))))))..(((....)--))...----)).))))) ( -20.90) >DroYak_CAF1 10103 96 - 1 -------------ACUCC----CUUCU----CAAUUUCUCGGCCAAGUCCAUCUCAUUUACAAUGGUCACCACUGAGAAAGCGAACUUGGCCAUCUGAAA---GAUUUCAUUGACAAGAA -------------.....----(((.(----((((.....((((((((...((.(.(((.((.(((...))).)).))).).))))))))))....(((.---...)))))))).))).. ( -21.80) >DroAna_CAF1 11108 112 - 1 UUUUCGGGAGCUCUCUAG----UUUUU----AAAUUUUUCCAUUAGGUCCAUUCCCUGAACGAGUGUAAAGACUGAAAAGGCAAACUUGACCAUUUGUGGAAUAAAAUAAUGUAGAACAC .....(((....)))..(----((((.----..(((.((((((..(((((((((.......)))))......((....))........))))....))))))......)))..))))).. ( -18.70) >consensus _____________ACUCC____CUUCU____CAAUUUCUCGGCCAAGUCCAUCUCAUUUACAAUGGUCACCACUGAGAAAGCGAACUUGGCCAUCUGAAA___GAUUUCAUUAACAAUAA ........................................((((((((...((.(.(((.((.((.....)).)).))).).))))))))))............................ (-11.76 = -11.32 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:32 2006