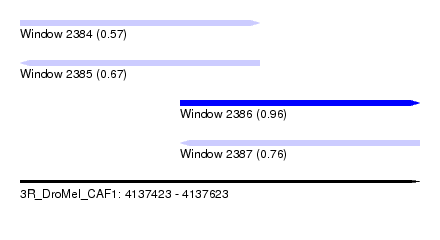
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,137,423 – 4,137,623 |
| Length | 200 |
| Max. P | 0.963225 |

| Location | 4,137,423 – 4,137,543 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -48.54 |
| Consensus MFE | -33.51 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
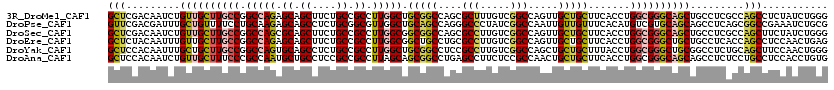
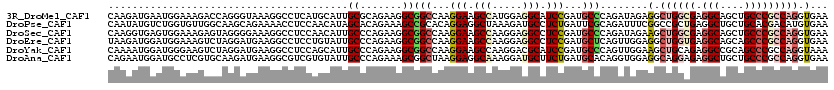
>3R_DroMel_CAF1 4137423 120 + 27905053 GCUCGACAAUCUGUUGCUUGCCGGCCAGAGCAGCUUCUGCCGCCUUGGCUGCGGCCAGCGCUUUGUCGGCCAGUUGCUGCUUCACCUGGCGGGCAGCUGCCUCGCCAGCCUCUAUCUGGG ...(((......((((((((((((...(((((((..((((((.(..(((.((.....)))))..).))).)))..)))))))...))))))))))))....)))((((.......)))). ( -52.20) >DroPse_CAF1 7526 120 + 1 GUUCGACGAUUUGCUGUUUUCCUGCAAGAGCAGCCUCUGCGGCGUUGGCUGCAGCCAGGGCCCUAUCGGCCAAUUGUUGUUUCACAUGUCGUGCAGCAGCCUCAGCGGCCGAAAUCUGCG ............((((((((......))))))))....(((((.(((((....))))).)))...((((((....((((((.(((.....))).))))))......)))))).....)). ( -44.00) >DroSec_CAF1 4191 120 + 1 GCUCGACAAUCUGUUGCUUGCCGGCCAGCGCAGCUUCUGCCGCCUUGGCGGCGGCCAGCGCCUUGUCGGCCAGUUGCUGCUUCACCUGGCGGGCAGCUGCCUCGCCAGCUUCUAUCUGGG ...(((......((((((((((((..(((((((((...(((((....)))))((((.((.....)).)))))))))).)))....))))))))))))....)))((((.......)))). ( -55.00) >DroEre_CAF1 4685 120 + 1 GCUCUACAAUUUGUUGCUUGCCGGCCAGAGCAGCUUCUGCCGCCUUGGCGGCUGCCUGCGCCUUGUCGGCCAGUUGCUGCUUCACCUGGCGGGCUGCUGCCUCACCAGCCUCCAACUGAG .(((...........(((((((((...(((((((....(((((....)))))...(((.(((.....))))))..)))))))...))))))))).((((......))))........))) ( -50.20) >DroYak_CAF1 8039 120 + 1 GCUCCACAAUUUGCUGCUUGCCGGCCAGUGCAGCCUCUGCCGCCUUGGCUGCGGCCUCCGCCUUGUCGGCCAGCUGCUGCUUUACCUGGCGGGCUGCGGCCUCUGCAGCUUCCAACUGGG ...(((......(((((.....((((...(((((((..((((....(((.(((((....(((.....)))..))))).))).....)))))))))))))))...))))).......))). ( -51.22) >DroAna_CAF1 4019 120 + 1 GCUCCACAAUCUGUUGCUUUCCCGCCAAUGCUGCCUCCGCCGCCUUAGCAGCGGCCUGAGCCUUCUCCGCCAACUGCUGCUUCACCUGGCGGGCAGCAGCCUCUCCUGCCUCCACCUGUG ....((((....(..((......((...(((((((..(((((....((((((((..((.((.......)))).)))))))).....)))))))))))))).......))..)....)))) ( -38.62) >consensus GCUCGACAAUCUGUUGCUUGCCGGCCAGAGCAGCCUCUGCCGCCUUGGCUGCGGCCAGCGCCUUGUCGGCCAGUUGCUGCUUCACCUGGCGGGCAGCUGCCUCACCAGCCUCCAUCUGGG (((.........((((((((((.(((((.((.((....)).)).))))).(((((....(((.....))).....))))).......)))))))))).........)))........... (-33.51 = -34.10 + 0.59)
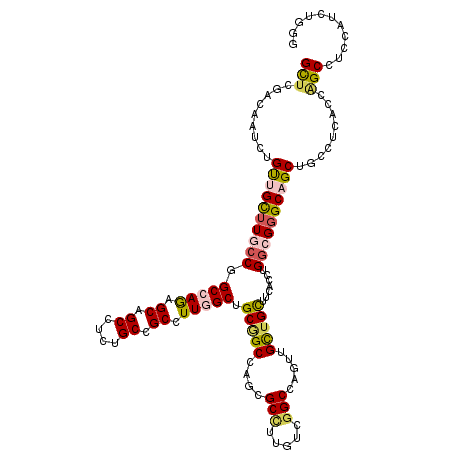
| Location | 4,137,423 – 4,137,543 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -52.60 |
| Consensus MFE | -31.33 |
| Energy contribution | -33.50 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137423 120 - 27905053 CCCAGAUAGAGGCUGGCGAGGCAGCUGCCCGCCAGGUGAAGCAGCAACUGGCCGACAAAGCGCUGGCCGCAGCCAAGGCGGCAGAAGCUGCUCUGGCCGGCAAGCAACAGAUUGUCGAGC .((((.......((((((.(((....)))))))))((......))..)))).((((((...(((.((((..((((..(((((....)))))..)))))))).)))......))))))... ( -55.30) >DroPse_CAF1 7526 120 - 1 CGCAGAUUUCGGCCGCUGAGGCUGCUGCACGACAUGUGAAACAACAAUUGGCCGAUAGGGCCCUGGCUGCAGCCAACGCCGCAGAGGCUGCUCUUGCAGGAAAACAGCAAAUCGUCGAAC .......((((((.((((.((((((.((......(((......)))...((((.....))))...)).))))))....((((((((....))).))).))....)))).....)))))). ( -41.80) >DroSec_CAF1 4191 120 - 1 CCCAGAUAGAAGCUGGCGAGGCAGCUGCCCGCCAGGUGAAGCAGCAACUGGCCGACAAGGCGCUGGCCGCCGCCAAGGCGGCAGAAGCUGCGCUGGCCGGCAAGCAACAGAUUGUCGAGC .((((.......((((((.(((....))))))))).....(((((..(((((((.(.....).)))))(((((....)))))))..))))).)))).((((((........))))))... ( -58.50) >DroEre_CAF1 4685 120 - 1 CUCAGUUGGAGGCUGGUGAGGCAGCAGCCCGCCAGGUGAAGCAGCAACUGGCCGACAAGGCGCAGGCAGCCGCCAAGGCGGCAGAAGCUGCUCUGGCCGGCAAGCAACAAAUUGUAGAGC ....((((...((((((.(((((((.(((.(((((.((......)).)))(((.....))))).))).(((((....)))))....))))).)).))))))...))))............ ( -56.70) >DroYak_CAF1 8039 120 - 1 CCCAGUUGGAAGCUGCAGAGGCCGCAGCCCGCCAGGUAAAGCAGCAGCUGGCCGACAAGGCGGAGGCCGCAGCCAAGGCGGCAGAGGCUGCACUGGCCGGCAAGCAGCAAAUUGUGGAGC (((((((....(((((...((((.....(((((.(((..(((....))).))).....))))).))))((.((((..(((((....)))))..))))..))..))))).))))).))... ( -54.60) >DroAna_CAF1 4019 120 - 1 CACAGGUGGAGGCAGGAGAGGCUGCUGCCCGCCAGGUGAAGCAGCAGUUGGCGGAGAAGGCUCAGGCCGCUGCUAAGGCGGCGGAGGCAGCAUUGGCGGGAAAGCAACAGAUUGUGGAGC (((((.((..(((((......))))).((((((((((......)).((((.(.(((....)))...(((((((....))))))).).)))).)))))))).......))..))))).... ( -48.70) >consensus CCCAGAUGGAGGCUGGAGAGGCAGCUGCCCGCCAGGUGAAGCAGCAACUGGCCGACAAGGCGCAGGCCGCAGCCAAGGCGGCAGAAGCUGCUCUGGCCGGCAAGCAACAAAUUGUCGAGC ...........((((((.((((((((..(((((..((......))...((((.(....(((....))).).)))).)))))....)))))).)).))))))................... (-31.33 = -33.50 + 2.17)

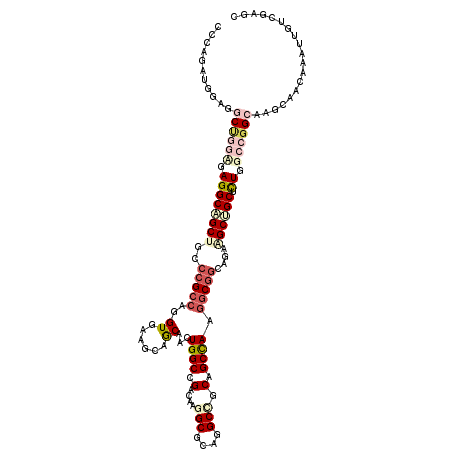

| Location | 4,137,503 – 4,137,623 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -20.79 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137503 120 + 27905053 UUCACCUGGCGGGCAGCUGCCUCGCCAGCCUCUAUCUGGGCAUCGGAUGCCUCCAUGGCUUCCUUGGCCGCCUUCUGCGCAAUGCAUGAGGCCUUUACCCUGGUCUUUCCAUUCAUCUUG .....(((((((((....))).))))))..........(((...(((.(((.....))).)))...)))(((((.(((.....))).)))))........(((.....)))......... ( -43.60) >DroPse_CAF1 7606 120 + 1 UUCACAUGUCGUGCAGCAGCCUCAGCGGCCGAAAUCUGCGAAUCAGAGGCAUCUUUAGCCUCCUGUGCGGCUUUCUGUGCUAUGUUGGAGGUUUUCUGCUUGCCAACACCAGACAUAUUG .....((((((..(((.((((.((((((.......))))....(((((((.......)))).))))).))))..)))..)..((((((((((.....)))).))))))...))))).... ( -41.20) >DroSec_CAF1 4271 120 + 1 UUCACCUGGCGGGCAGCUGCCUCGCCAGCUUCUAUCUGGGCAUCGGAGGCCUCCUUGGCUUCCUUGGCCGCCUUCUGGGCAAUGUUGGAGGCCUUUCCCCUACUCUUUCCACUCACCUUG .....(((((((((....))).)))))).........(((....((((((((((..((((.....))))(((.....)))......)))))))))).))).................... ( -49.10) >DroEre_CAF1 4765 120 + 1 UUCACCUGGCGGGCUGCUGCCUCACCAGCCUCCAACUGAGCAUCGGAGGCCUCCUUGGCUUCCUUGGCCGCCUUCUGGGCAAUACAGGAGGCCUUCAUCCUAGACUUUCCAUCCAUCUUA ((((..(((.((((((.((...)).)))))))))..))))....(((((((((((.((((.....))))(((.....))).....)))))))))))........................ ( -46.90) >DroYak_CAF1 8119 120 + 1 UUUACCUGGCGGGCUGCGGCCUCUGCAGCUUCCAACUGGGCAUCGGAUGCGUCCUUGGCUUCCUUGGCCGCCUUCUGGGCAAUGCUGGAGGCCUUCAUCCUAGACUUCCCAUCCAUUUUG ......(((.(((((((((...))))))))))))..((((..(((((((.......(((((((..(((.(((.....)))...))))))))))..)))))..))...))))......... ( -42.60) >DroAna_CAF1 4099 120 + 1 UUCACCUGGCGGGCAGCAGCCUCUCCUGCCUCCACCUGUGCAUCAGAAGCAUCCUUUGCCUCCUUAGCCGCUUUCUGGGCAAUACACGACGCCUUCAUCUUGCACGAGGCAUCCAUUCUG ......(((.((((((.((...)).)))))))))((((((((...(((((.((..((((((....((...))....)))))).....)).).))))....))))).)))........... ( -31.00) >consensus UUCACCUGGCGGGCAGCUGCCUCACCAGCCUCCAUCUGGGCAUCGGAGGCCUCCUUGGCUUCCUUGGCCGCCUUCUGGGCAAUGCAGGAGGCCUUCAUCCUACACUUUCCAUCCAUCUUG ......(((.((((....))).).)))(((((......(((...(((.(((.....))).)))...)))(((.....))).......)))))............................ (-20.79 = -22.23 + 1.45)
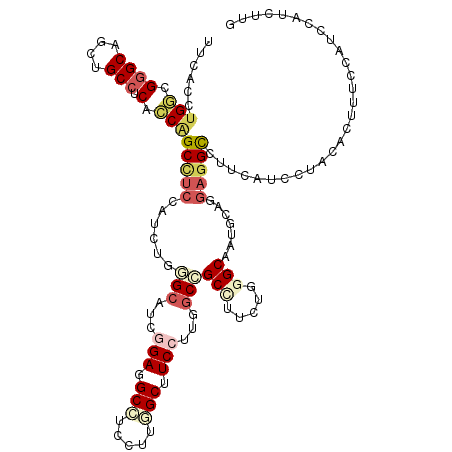
| Location | 4,137,503 – 4,137,623 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137503 120 - 27905053 CAAGAUGAAUGGAAAGACCAGGGUAAAGGCCUCAUGCAUUGCGCAGAAGGCGGCCAAGGAAGCCAUGGAGGCAUCCGAUGCCCAGAUAGAGGCUGGCGAGGCAGCUGCCCGCCAGGUGAA .........(((.....)))(((((..((((.(.(((.....)))....).))))..(((.(((.....))).)))..))))).........((((((.(((....)))))))))..... ( -47.30) >DroPse_CAF1 7606 120 - 1 CAAUAUGUCUGGUGUUGGCAAGCAGAAAACCUCCAACAUAGCACAGAAAGCCGCACAGGAGGCUAAAGAUGCCUCUGAUUCGCAGAUUUCGGCCGCUGAGGCUGCUGCACGACAUGUGAA ..((((((((((((((((..((........)))))))))(((...((((.(.((...((((((.......)))))).....)).).))))((((.....))))))).)).)))))))... ( -37.90) >DroSec_CAF1 4271 120 - 1 CAAGGUGAGUGGAAAGAGUAGGGGAAAGGCCUCCAACAUUGCCCAGAAGGCGGCCAAGGAAGCCAAGGAGGCCUCCGAUGCCCAGAUAGAAGCUGGCGAGGCAGCUGCCCGCCAGGUGAA .........(((.....(((..(((..(((((((......(((.....)))(((.......)))..))))))))))..))))))........((((((.(((....)))))))))..... ( -47.20) >DroEre_CAF1 4765 120 - 1 UAAGAUGGAUGGAAAGUCUAGGAUGAAGGCCUCCUGUAUUGCCCAGAAGGCGGCCAAGGAAGCCAAGGAGGCCUCCGAUGCUCAGUUGGAGGCUGGUGAGGCAGCAGCCCGCCAGGUGAA .....(((((.....))))).......((((((((.....(((.....)))(((.......))).))))))))((((((.....))))))..((((((.(((....)))))))))..... ( -47.90) >DroYak_CAF1 8119 120 - 1 CAAAAUGGAUGGGAAGUCUAGGAUGAAGGCCUCCAGCAUUGCCCAGAAGGCGGCCAAGGAAGCCAAGGACGCAUCCGAUGCCCAGUUGGAAGCUGCAGAGGCCGCAGCCCGCCAGGUAAA .....(((.((((...((......)).((((((((((...(((.....)))(((...(((.((.......)).)))...))).........))))..))))))....)))))))...... ( -41.10) >DroAna_CAF1 4099 120 - 1 CAGAAUGGAUGCCUCGUGCAAGAUGAAGGCGUCGUGUAUUGCCCAGAAAGCGGCUAAGGAGGCAAAGGAUGCUUCUGAUGCACAGGUGGAGGCAGGAGAGGCUGCUGCCCGCCAGGUGAA .......((((((((((.....))).)))))))(((((((((((.....).)))...(((((((.....)))))))))))))).(((((.(((((......)))))..)))))....... ( -48.10) >consensus CAAGAUGGAUGGAAAGUCCAGGAUGAAGGCCUCCAGCAUUGCCCAGAAGGCGGCCAAGGAAGCCAAGGAGGCCUCCGAUGCCCAGAUGGAGGCUGGAGAGGCAGCUGCCCGCCAGGUGAA ........................................((.......))(((...(((.(((.....))).)))...)))........(.((((((.(((....))))))))).)... (-19.91 = -20.88 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:26 2006