
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,137,026 – 4,137,223 |
| Length | 197 |
| Max. P | 0.937060 |

| Location | 4,137,026 – 4,137,143 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -28.33 |
| Energy contribution | -29.67 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
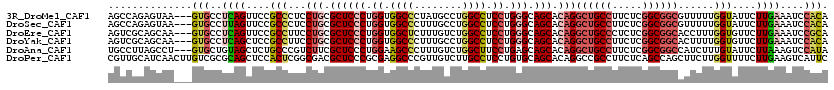
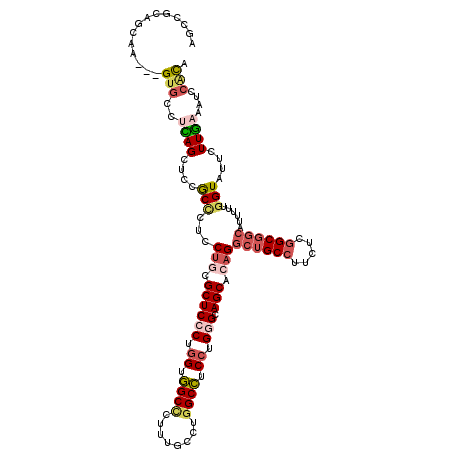
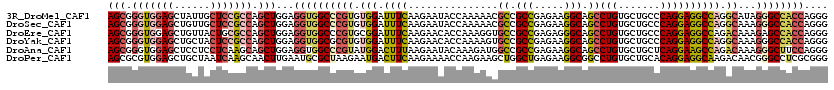
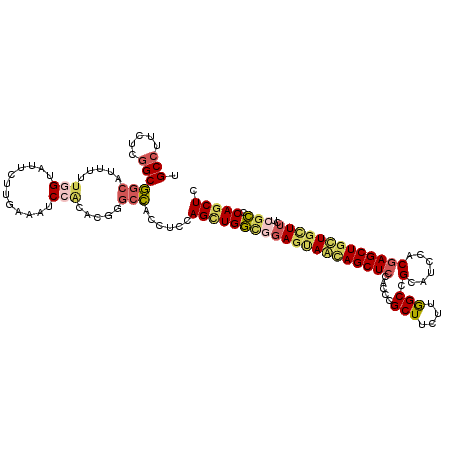
>3R_DroMel_CAF1 4137026 117 + 27905053 AGCCAGAGUAA---GUGCCUCAGUUCCGCCCUCCUGCGCUCCCUGGUGGCCCUAUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACA .(((((((...---....))).....((((..((((.((((((.((.((((........)))).)).))).))).))))..(((.....)))))))....))))................ ( -44.00) >DroSec_CAF1 3794 117 + 1 AGCCAGAGUAA---GUGCCUUAGUUCCGCCCUCCUGCGCUCCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACA .....(((..(---(((((..((...((((..((((.((((((.((.((((........)))).)).))).))).))))..(((.....))))))).))..))))))))).......... ( -44.30) >DroEre_CAF1 4288 117 + 1 AGUCGCAGCAA---GUGCCUCAGUUCCGCCUUCCUGCGCUCCCUGGUGGCUCUUUGUCUGGCCUCCUGGGCAGCACAGGCUGCCCUCUCGGCGGCACCUUUGGUGUUCUUGAAAUCCGCA ....((.((..---..)).((((...((((...(((.((((((.((.((((........)))).)).))).))).)))((((((.....))))))......))))...)))).....)). ( -43.90) >DroYak_CAF1 7642 117 + 1 AGUCGCAGCAA---GUGCCUCAGCUCCGCCUUCCUGCGCUCCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCACUUUUGGUGUUCUUGAAAUCCACA ...(((...((---(((((.............((((.((((((.((.((((........)))).)).))).))).))))..(((.....))))))))))...)))............... ( -45.60) >DroAna_CAF1 3622 117 + 1 UGCCUUAGCCU---GUGCUGUAGCUCUGCCCGUCUUCGCUCCCUGGAAGCCCUUUGUCUGGCUUCCUGAGCAGCACAGGCUGCCUUCUCGGCGGCCAUCUUUGUAUUCUUAAAGUCCAUA .(((..(((((---((((((((....)))........((((...(((((((........))))))).))))))))))))))(((.....))))))...(((((......)))))...... ( -47.40) >DroPer_CAF1 9640 120 + 1 CGUUGCAUCAACUUGUCGCGCAGCUCCACUCGGCGACGCUCCCGCGAGGCCCGUUGUCUUGCCUCCUGUGCAGCACAGGCCGCCUUCUCAGCCAGCUUCUUGGUUUUCUUGAAGUCAUUC .(((((.............))))).......(((((((...((....))..)))))))..((..((((((...))))))..))((((..((((((....)))))).....))))...... ( -32.92) >consensus AGCCGCAGCAA___GUGCCUCAGCUCCGCCCUCCUGCGCUCCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCAUUUUUGGUAUUCUUGAAAUCCACA ..............(((..((((....(((...(((.((((((.((.((((........)))).)).))).))).)))((((((.....))))))......)))....))))....))). (-28.33 = -29.67 + 1.34)
| Location | 4,137,026 – 4,137,143 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -47.01 |
| Consensus MFE | -31.02 |
| Energy contribution | -32.13 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
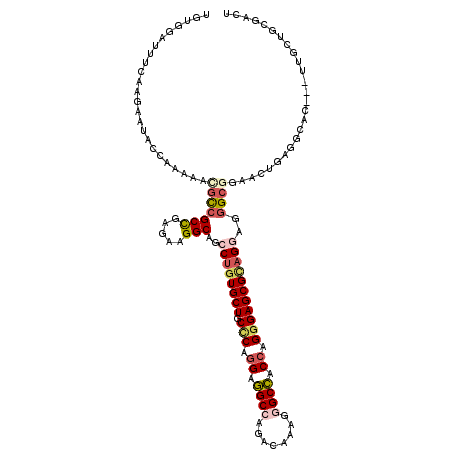
>3R_DroMel_CAF1 4137026 117 - 27905053 UGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAUAGGGCCACCAGGGAGCGCAGGAGGGCGGAACUGAGGCAC---UUACUCUGGCU .................(((....(((((((.....)))..((((((((.(((.((.((((........)))).)).)))))))))))..)))).....(((....---...)))))).. ( -48.60) >DroSec_CAF1 3794 117 - 1 UGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAAAGGGCCACCAGGGAGCGCAGGAGGGCGGAACUAAGGCAC---UUACUCUGGCU ..(((((((...)))).)))....(((((((.....)))..((((((((.(((.((.((((........)))).)).)))))))))))..)))).......(((.(---.......)))) ( -46.80) >DroEre_CAF1 4288 117 - 1 UGCGGAUUUCAAGAACACCAAAGGUGCCGCCGAGAGGGCAGCCUGUGCUGCCCAGGAGGCCAGACAAAGAGCCACCAGGGAGCGCAGGAAGGCGGAACUGAGGCAC---UUGCUGCGACU .((((................(((((((.((....))((..((((((((.(((.((.((((.......).))).)).)))))))))))...))........)))))---)).)))).... ( -49.13) >DroYak_CAF1 7642 117 - 1 UGUGGAUUUCAAGAACACCAAAAGUGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAAAGGGCCACCAGGGAGCGCAGGAAGGCGGAGCUGAGGCAC---UUGCUGCGACU .(((...........)))((.((((((((((.....)))..((((((((.(((.((.((((........)))).)).))))))))))).............)))))---))..))..... ( -49.20) >DroAna_CAF1 3622 117 - 1 UAUGGACUUUAAGAAUACAAAGAUGGCCGCCGAGAAGGCAGCCUGUGCUGCUCAGGAAGCCAGACAAAGGGCUUCCAGGGAGCGAAGACGGGCAGAGCUACAGCAC---AGGCUAAGGCA ......((((........))))...((((((.....)))((((((((((((((.(((((((........))))))).....((........)).))))...)))))---)))))..))). ( -50.40) >DroPer_CAF1 9640 120 - 1 GAAUGACUUCAAGAAAACCAAGAAGCUGGCUGAGAAGGCGGCCUGUGCUGCACAGGAGGCAAGACAACGGGCCUCGCGGGAGCGUCGCCGAGUGGAGCUGCGCGACAAGUUGAUGCAACG ......(((((......(((......))).......(((((((((((...)))))(((((..(....)..)))))((....))))))))...))))).(((((((....))).))))... ( -37.90) >consensus UGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGACAAAGGGCCACCAGGGAGCGCAGGAGGGCGGAACUGAGGCAC___UUGCUGCGACU ........................(((((((.....)))..((((((((.(((.((.((((........)))).)).)))))))))))..)))).......................... (-31.02 = -32.13 + 1.12)


| Location | 4,137,063 – 4,137,183 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -49.98 |
| Consensus MFE | -33.94 |
| Energy contribution | -36.30 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137063 120 + 27905053 CCCUGGUGGCCCUAUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGCAAUAGCUCCACCCGCU ....((((((((.(((((..(((....(((((((....)))))))....))))))))...(((.(((.....))))))...))))))))...(((.((.(((((....))))).)).))) ( -58.60) >DroSec_CAF1 3831 120 + 1 CCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGCAACAGCUCCACCCGCU ....((((((((...(((..(((....(((((((....)))))))....)))))).....(((.(((.....))))))...))))))))...(((.((.(((((....))))).)).))) ( -57.70) >DroEre_CAF1 4325 120 + 1 CCCUGGUGGCUCUUUGUCUGGCCUCCUGGGCAGCACAGGCUGCCCUCUCGGCGGCACCUUUGGUGUUCUUGAAAUCCGCACGGGCCACCUCCAGCUGGCGCAGUAACAGCUCCACCCGCU ....((((((((..(((...(((....(((((((....)))))))....)))((((((...))))))..........))).))))))))...(((.((.(.(((....))).).)).))) ( -48.00) >DroYak_CAF1 7679 120 + 1 CCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCACUUUUGGUGUUCUUGAAAUCCACACGCGCCACCUCCAGCUGGCGGAGUAGCAGCUCCACCCGCU ....((((((....((((..(((....(((((((....)))))))....)))))))......((((...........))))..))))))...(((.((.(((((....))))).)).))) ( -50.30) >DroAna_CAF1 3659 120 + 1 CCCUGGAAGCCCUUUGUCUGGCUUCCUGAGCAGCACAGGCUGCCUUCUCGGCGGCCAUCUUUGUAUUCUUAAAGUCCAUACGGGCCACCUCCAGCUGCUUGAGGAGGAGCUCCACCCGCU .((((((((((........))))))).(((((((...(((((((.....))))))).................((((....))))........))))))).))).((....))....... ( -47.20) >DroPer_CAF1 9680 120 + 1 CCCGCGAGGCCCGUUGUCUUGCCUCCUGUGCAGCACAGGCCGCCUUCUCAGCCAGCUUCUUGGUUUUCUUGAAGUCAUUCUUAGCGCAUUCAAGUUGCUUGAUUAGCAGCUCCACGCGCU .(((.(((((..((((....((..((((((...))))))..)).....))))..))))).)))...................(((((.....(((((((.....)))))))....))))) ( -38.10) >consensus CCCUGGUGGCCCUUUGCCUGGCCUCCUGGGCAGCACAGGCUGCCUUCUCGGCGGCAUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGUAACAGCUCCACCCGCU ....((((((((...(((..(((....(((((((....)))))))....))))))...(((.(......).))).......))))))))...(((.((.(((((....))))).)).))) (-33.94 = -36.30 + 2.36)


| Location | 4,137,063 – 4,137,183 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -53.53 |
| Consensus MFE | -35.27 |
| Energy contribution | -36.75 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137063 120 - 27905053 AGCGGGUGGAGCUAUUGCUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAUAGGGCCACCAGGG (((.((((((((....)))))))).)))...((((((((((.(((((((...)))).)))...))((((((.....((((((....)))))).....)))..)))...)))))))).... ( -64.30) >DroSec_CAF1 3831 120 - 1 AGCGGGUGGAGCUGUUGCUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAAAGGGCCACCAGGG (((.((((((((....)))))))).)))...((((((((((.(((((((...)))).)))...))((((((.....((((((....)))))).....)))..)))...)))))))).... ( -64.30) >DroEre_CAF1 4325 120 - 1 AGCGGGUGGAGCUGUUACUGCGCCAGCUGGAGGUGGCCCGUGCGGAUUUCAAGAACACCAAAGGUGCCGCCGAGAGGGCAGCCUGUGCUGCCCAGGAGGCCAGACAAAGAGCCACCAGGG ....(((((..((....(((.(((.......((((((((....((............))...)).))))))....(((((((....)))))))....))))))....))..))))).... ( -51.20) >DroYak_CAF1 7679 120 - 1 AGCGGGUGGAGCUGCUACUCCGCCAGCUGGAGGUGGCGCGUGUGGAUUUCAAGAACACCAAAAGUGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGGCAAAGGGCCACCAGGG (((.(((((((......))))))).)))...((((((((...(((............)))...)))))))).....((((((....))))))..((.((((........)))).)).... ( -56.10) >DroAna_CAF1 3659 120 - 1 AGCGGGUGGAGCUCCUCCUCAAGCAGCUGGAGGUGGCCCGUAUGGACUUUAAGAAUACAAAGAUGGCCGCCGAGAAGGCAGCCUGUGCUGCUCAGGAAGCCAGACAAAGGGCUUCCAGGG ...(((.((....)).)))..((((((.(.((((((((.((((...........))))......))))(((.....))).)))).)))))))..(((((((........))))))).... ( -45.00) >DroPer_CAF1 9680 120 - 1 AGCGCGUGGAGCUGCUAAUCAAGCAACUUGAAUGCGCUAAGAAUGACUUCAAGAAAACCAAGAAGCUGGCUGAGAAGGCGGCCUGUGCUGCACAGGAGGCAAGACAACGGGCCUCGCGGG (((((((.(((.((((.....)))).)))..)))))))...................((.....((((.((....)).))))(((((...)))))(((((..(....)..)))))..)). ( -40.30) >consensus AGCGGGUGGAGCUGCUACUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCAGCCUGUGCUGCCCAGGAGGCCAGACAAAGGGCCACCAGGG (((.(((((((......))))))).)))...((((((((((.(((.((((...............((.(((.....))).))(((.......)))))))))).))...)))))))).... (-35.27 = -36.75 + 1.48)


| Location | 4,137,103 – 4,137,223 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -29.69 |
| Energy contribution | -30.45 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137103 120 + 27905053 UGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGCAAUAGCUCCACCCGCUUCUUGGCCGCUUCCACAAGCUGUUGCUUCUCGCCCAGCUC .(((.....)))((((.....((((.................(((((.....(((.((.(((((....))))).)).)))...)))))((((....))))...))))...))))...... ( -40.50) >DroPse_CAF1 7206 120 + 1 CGCCUUCUCAGCCAGCUUCUUGGUUUUCUUGAAGUCAUUCUUAGCGCAUUCAAGUUGCUUGAUUAGCAGCUCCACGCGCUUCUUAGCGGCCUCCACGAGCUGCUGCUUCUCGGACAGCUC (((((((..((((((....)))))).....))))........(((((.....(((((((.....)))))))....))))).....)))........(((((((((.....))).)))))) ( -35.70) >DroSec_CAF1 3871 120 + 1 UGCCUUCUCGGCGGCGUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGCAACAGCUCCACCCGCUUCUUGGCCGCCUCCACGAGCUGUUGCUUCUCGCCCAGCUC .(((.....)))(((.....(((.(((.....))))))....(((((.....(((.((.(((((....))))).)).)))...)))))))).....((((((..((.....)).)))))) ( -45.00) >DroEre_CAF1 4365 120 + 1 UGCCCUCUCGGCGGCACCUUUGGUGUUCUUGAAAUCCGCACGGGCCACCUCCAGCUGGCGCAGUAACAGCUCCACCCGCUUCUUGGCCGCAACCACGAGCUGCUGCUUCUCGCUCAACUC (((.....((((((((((...))))..........)))).))(((((.....(((.((.(.(((....))).).)).)))...)))))))).....((((...........))))..... ( -32.70) >DroYak_CAF1 7719 120 + 1 UGCCUUCUCGGCGGCACUUUUGGUGUUCUUGAAAUCCACACGCGCCACCUCCAGCUGGCGGAGUAGCAGCUCCACCCGCUUCUUGGCCGCAUCCACGAGCUGCUGCUUCUCGCCCAGCUC .(((.....)))((......((((((...((.....))...))))))...))((((((((((((((((((((.....(((....))).(......))))))))))).)).)).)))))). ( -43.50) >DroAna_CAF1 3699 120 + 1 UGCCUUCUCGGCGGCCAUCUUUGUAUUCUUAAAGUCCAUACGGGCCACCUCCAGCUGCUUGAGGAGGAGCUCCACCCGCUUCUUGGCAGCUUCCACGAGCUGCUGUUUCUCCGACAGCUC .(((.....)))((((..(((((......)))))........))))......(((((...((((((((((.......)))))))((((((((....))))))))....)))...))))). ( -41.20) >consensus UGCCUUCUCGGCGGCAUUUUUGGUAUUCUUGAAAUCCACACGGGCCACCUCCAGCUGGCGGAGUAACAGCUCCACCCGCUUCUUGGCCGCAUCCACGAGCUGCUGCUUCUCGCCCAGCUC .(((.....)))(((.....(((............))).....)))......((((((((((((((((((((.....(((....))).(......)))))))))))))..))).))))). (-29.69 = -30.45 + 0.76)


| Location | 4,137,103 – 4,137,223 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -31.43 |
| Energy contribution | -32.60 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4137103 120 - 27905053 GAGCUGGGCGAGAAGCAACAGCUUGUGGAAGCGGCCAAGAAGCGGGUGGAGCUAUUGCUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCA ((((((.((.....))..))))))..((....(((((...(((.((((((((....)))))))).))).....)))))(((.(((((((...)))).)))...)))))(((.....))). ( -51.60) >DroPse_CAF1 7206 120 - 1 GAGCUGUCCGAGAAGCAGCAGCUCGUGGAGGCCGCUAAGAAGCGCGUGGAGCUGCUAAUCAAGCAACUUGAAUGCGCUAAGAAUGACUUCAAGAAAACCAAGAAGCUGGCUGAGAAGGCG ..(((((.......))))).(((..(...(((((((....(((((((.(((.((((.....)))).)))..)))))))..(((....))).............))).))))..)..))). ( -36.90) >DroSec_CAF1 3871 120 - 1 GAGCUGGGCGAGAAGCAACAGCUCGUGGAGGCGGCCAAGAAGCGGGUGGAGCUGUUGCUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCA ((((((.((.....))..)))))).....((((((((...(((.((((((((....)))))))).))).....))))).((.(((((((...)))).)))...)))))(((.....))). ( -57.40) >DroEre_CAF1 4365 120 - 1 GAGUUGAGCGAGAAGCAGCAGCUCGUGGUUGCGGCCAAGAAGCGGGUGGAGCUGUUACUGCGCCAGCUGGAGGUGGCCCGUGCGGAUUUCAAGAACACCAAAGGUGCCGCCGAGAGGGCA .(((((.(((((.((((((.(((((((((....))).....))))))...)))))).)).)))))))).......((((..((((..........((((...)))))))).....)))). ( -43.70) >DroYak_CAF1 7719 120 - 1 GAGCUGGGCGAGAAGCAGCAGCUCGUGGAUGCGGCCAAGAAGCGGGUGGAGCUGCUACUCCGCCAGCUGGAGGUGGCGCGUGUGGAUUUCAAGAACACCAAAAGUGCCGCCGAGAAGGCA ((((((.((.....))..)))))).........(((....(((.(((((((......))))))).)))...((((((((...(((............)))...)))))))).....))). ( -54.00) >DroAna_CAF1 3699 120 - 1 GAGCUGUCGGAGAAACAGCAGCUCGUGGAAGCUGCCAAGAAGCGGGUGGAGCUCCUCCUCAAGCAGCUGGAGGUGGCCCGUAUGGACUUUAAGAAUACAAAGAUGGCCGCCGAGAAGGCA (((((((.(......).))))))).........((((....((((((.(..((((..((.....))..)))).).)))))).....((((........)))).)))).(((.....))). ( -40.70) >consensus GAGCUGGGCGAGAAGCAGCAGCUCGUGGAAGCGGCCAAGAAGCGGGUGGAGCUGCUACUCCGCCAGCUGGAGGUGGCCCGUGUGGAUUUCAAGAAUACCAAAAACGCCGCCGAGAAGGCA ((((((............)))))).........(((....(((.(((((((......))))))).)))...((((((.....(((............))).....)))))).....))). (-31.43 = -32.60 + 1.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:22 2006