
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,130,403 – 4,130,563 |
| Length | 160 |
| Max. P | 0.901734 |

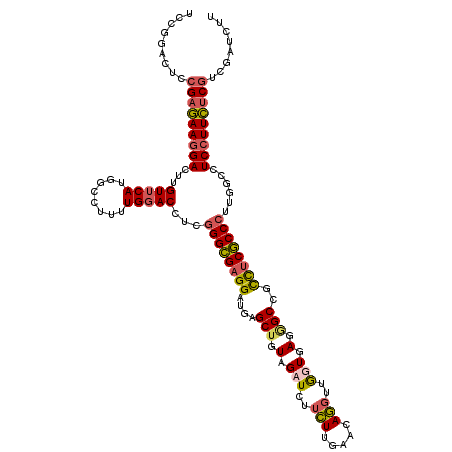
| Location | 4,130,403 – 4,130,523 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -43.96 |
| Consensus MFE | -29.31 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4130403 120 + 27905053 UCCGGACUCCGAGAAGGACUUGUUCAUGGCCUUUUGGACCUCGGGCGAGGAUGAGCUGUAGAUUUUCUUGAACAGGUUGGUGAGGGCCGCCUCGCCCUUGGCCUCCUUCUCGUCGAUCUU ...((((..(((((((((.........((((.....(((((.(..((((((....(....)...))))))..))))))(((((((....)))))))...)))))))))))))..).))). ( -45.30) >DroSim_CAF1 634 120 + 1 UCCCGACUCCGAGAAGGACUUGUUCAUGGCCUUUUGGACCUCGGGCGAGGAUGAGCUGUAGAUUUUCUUGAACAGGUUGGUGAGUGCCGCCUCGCCCUUGGCCUCCUUCUCGGCGAUCUU ....(((.((((((((((.........((((.....(....)((((((((..((.((((((.....))...)))).))(((....))).))))))))..)))))))))))))).).)).. ( -47.60) >DroWil_CAF1 1051 120 + 1 GCCGGACUCCGUAAAUGAUUUGUUCAUGGCCAUUUGCACCUCCGGUGUGGAGGUGCUGUAGAUCUUUUUGAAUAAUUGAUUGAGAGCCGCUUCACCUUUGGCUUCUUUUUCAUCGAUCUU (((((...))....((((.....))))))).....((((((((.....))))))))...(((((............(((..((((((((.........)))).))))..)))..))))). ( -32.94) >DroYak_CAF1 636 120 + 1 UCCGGAUUCCGAGAAGGACUUGUUCAUGGCCUUUUGGACCUCGGGCGAGGAUGUGCUGUAGAUCUUCUUGAACAGGUUGUUGAGGGCCGUCUCGCCCUUAGCCUCCUUCUCGUCGAUCUU ...(((((.(((((((((((((((((..((((..........))))((((((.((...)).)))))).))))))))..(((((((((......))))))))).)))))))))..))))). ( -50.00) >consensus UCCGGACUCCGAGAAGGACUUGUUCAUGGCCUUUUGGACCUCGGGCGAGGAUGAGCUGUAGAUCUUCUUGAACAGGUUGGUGAGGGCCGCCUCGCCCUUGGCCUCCUUCUCGUCGAUCUU .........(((((((((...(((((........)))))...((((((((....(((.(.(((..(((.....)))..))).).)))..))))))))......)))))))))........ (-29.31 = -30.00 + 0.69)

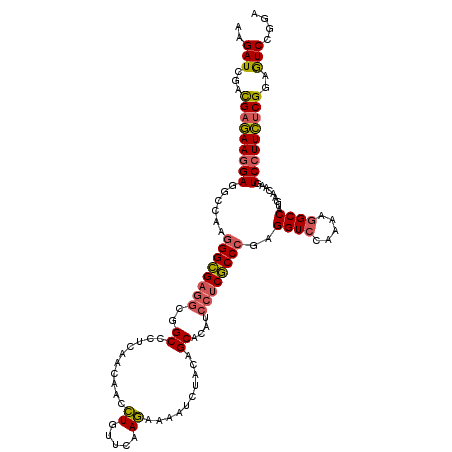
| Location | 4,130,403 – 4,130,523 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -29.35 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4130403 120 - 27905053 AAGAUCGACGAGAAGGAGGCCAAGGGCGAGGCGGCCCUCACCAACCUGUUCAAGAAAAUCUACAGCUCAUCCUCGCCCGAGGUCCAAAAGGCCAUGAACAAGUCCUUCUCGGAGUCCGGA ..((.(..(((((((((((((..((((((((.((......))...((((...((.....)))))).....))))))))..)))).....(........)...)))))))))..))).... ( -42.70) >DroSim_CAF1 634 120 - 1 AAGAUCGCCGAGAAGGAGGCCAAGGGCGAGGCGGCACUCACCAACCUGUUCAAGAAAAUCUACAGCUCAUCCUCGCCCGAGGUCCAAAAGGCCAUGAACAAGUCCUUCUCGGAGUCGGGA ..(((..((((((((((((((..((((((((.((......))...((((...((.....)))))).....))))))))..)))).....(........)...)))))))))).))).... ( -45.80) >DroWil_CAF1 1051 120 - 1 AAGAUCGAUGAAAAAGAAGCCAAAGGUGAAGCGGCUCUCAAUCAAUUAUUCAAAAAGAUCUACAGCACCUCCACACCGGAGGUGCAAAUGGCCAUGAACAAAUCAUUUACGGAGUCCGGC .(((((..((((.((((((((...........)))).))......)).))))....)))))...((((((((.....)))))))).....(((((((.....))))....((...))))) ( -29.30) >DroYak_CAF1 636 120 - 1 AAGAUCGACGAGAAGGAGGCUAAGGGCGAGACGGCCCUCAACAACCUGUUCAAGAAGAUCUACAGCACAUCCUCGCCCGAGGUCCAAAAGGCCAUGAACAAGUCCUUCUCGGAAUCCGGA ..(((...(((((((((......(((((((..(........)...((((...((.....))))))......)))))))..((((.....)))).........)))))))))..))).... ( -37.00) >consensus AAGAUCGACGAGAAGGAGGCCAAGGGCGAGGCGGCCCUCAACAACCUGUUCAAGAAAAUCUACAGCACAUCCUCGCCCGAGGUCCAAAAGGCCAUGAACAAGUCCUUCUCGGAGUCCGGA ..(((...(((((((((......((((((((..((..........((.....))..........))....))))))))..((((.....)))).........)))))))))..))).... (-29.35 = -29.60 + 0.25)


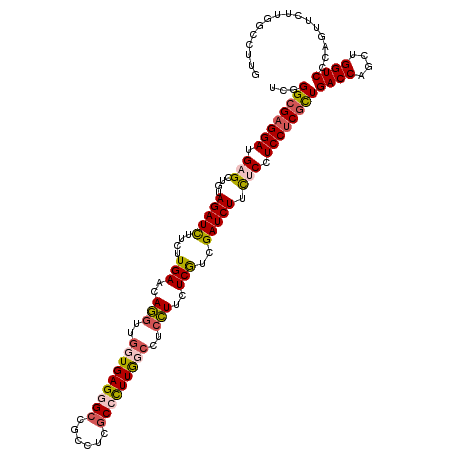
| Location | 4,130,443 – 4,130,563 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -39.71 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4130443 120 + 27905053 UCGGGCGAGGAUGAGCUGUAGAUUUUCUUGAACAGGUUGGUGAGGGCCGCCUCGCCCUUGGCCUCCUUCUCGUCGAUCUUUUCCUCCUCGCUGACCAGCUGGUCCCAGUUCUUGGCCUUG ..(((((((((.(((....(((((..(..(((.((((((((((((....)))))))...)))))..)))..)..))))).))).)))))))...(((((((....))))...)))))... ( -43.00) >DroSim_CAF1 674 120 + 1 UCGGGCGAGGAUGAGCUGUAGAUUUUCUUGAACAGGUUGGUGAGUGCCGCCUCGCCCUUGGCCUCCUUCUCGGCGAUCUUCUCCUCCUCGUUGACCAGCUGGUCCCAGUUUUUGGCCUUG ..((((((((..((.((((((.....))...)))).))(((....))).))))))))..((((......(((((((...........)))))))..(((((....)))))...))))... ( -42.20) >DroWil_CAF1 1091 120 + 1 UCCGGUGUGGAGGUGCUGUAGAUCUUUUUGAAUAAUUGAUUGAGAGCCGCUUCACCUUUGGCUUCUUUUUCAUCGAUCUUUUCCUCCUCCUUGACCAAAAGGUCCCAGUUCUUUGUUUGC ...((...(((((.(....(((((............(((..((((((((.........)))).))))..)))..)))))....).)))))..((((....)))))).............. ( -28.34) >DroYak_CAF1 676 120 + 1 UCGGGCGAGGAUGUGCUGUAGAUCUUCUUGAACAGGUUGUUGAGGGCCGUCUCGCCCUUAGCCUCCUUCUCGUCGAUCUUCUCCUCCUCGCUGACCAGCUGGUCCCAGUUCUUGGCCUUA ..(((((((((.(.(....(((((....(((..(((..(((((((((......)))))))))..)))..)))..))))).).).)))))))...(((((((....))))...)))))... ( -45.30) >consensus UCGGGCGAGGAUGAGCUGUAGAUCUUCUUGAACAGGUUGGUGAGGGCCGCCUCGCCCUUGGCCUCCUUCUCGUCGAUCUUCUCCUCCUCGCUGACCAGCUGGUCCCAGUUCUUGGCCUUG ...((((((((.(((....(((((....(((..(((..(((((((((......)))))))))..)))..)))..))))).))).))))))))((((....))))................ (-29.76 = -30.82 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:12 2006