
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,100,684 – 4,100,918 |
| Length | 234 |
| Max. P | 0.998064 |

| Location | 4,100,684 – 4,100,804 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -36.61 |
| Energy contribution | -36.45 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4100684 120 - 27905053 GCCGACUUUGAUGCGGCAAUUCGAGCGCGGUGGUGUUGUUUUGCCAACAUUACUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGUGAUCCUUUUAUCCUUCAU ((((.(......))))).....((((((((((((((((......)))))))))))).(((((.(......).)))))...)).((((.(((....)))))))..............)).. ( -39.00) >DroSec_CAF1 12542 120 - 1 GCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAAUGCUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGUGAUCCUUUUAUCCUUCAU ((((.(......))))).......(.((((..((((((((.....))))))))..)))).)......(((((.(((.......((((.(((....))))))).........)))))))). ( -38.09) >DroSim_CAF1 12605 120 - 1 GCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGUGAUCCUUUUAUCCUUCAU ...((..(((((.(((((...((....))(..((((((......))))))..)))))))))))..))(((((.(((.......((((.(((....))))))).........)))))))). ( -37.59) >DroEre_CAF1 12681 120 - 1 GCCGACUUUGAUGCGGCAAUUCGAACUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGUGAUCCCUUUAUCCCUCAU ...((..(((((.(((((...((....))(..((((((......))))))..)))))))))))..))(((((.(((.......((((.(((....))))))).........)))))))). ( -40.29) >DroYak_CAF1 12675 120 - 1 GCAGGCUUUGAUGCGGCAAUUCGAACUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGAGAUCCCCUUAUCCCUCAU .((((..(((((.(((((...((....))(..((((((......))))))..)))))))))))..))))(((.(((((......(((.(((....))))))))))).))).......... ( -40.40) >consensus GCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUCGCAACUGACUGGCGCCAGCAGUGAUCCUUUUAUCCUUCAU ...((..(((((.(((((...((....))(((((((((......)))))))))))))))))))..))(((((.(((.......((((.(((....))))))).........)))))))). (-36.61 = -36.45 + -0.16)

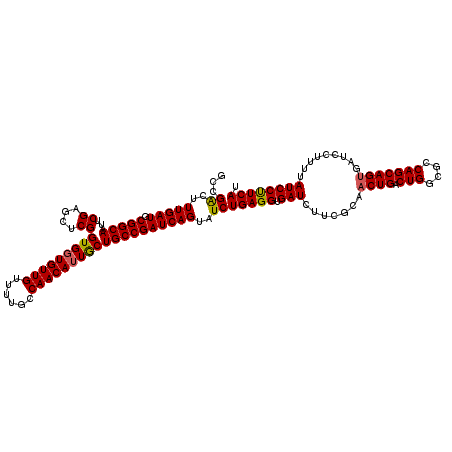

| Location | 4,100,724 – 4,100,844 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -29.00 |
| Energy contribution | -30.40 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4100724 120 + 27905053 GAAGAUCACCUCAGAUACUGAUCGGCAGUAAUGUUGGCAAAACAACACCACCGCGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGGUCCGAUCCAAUCUGGGCA .........(((((((...((((((.......((((......))))((((..((((.......).)))......((((((((......)))))..))).))))))))))..))))))).. ( -35.30) >DroSec_CAF1 12582 120 + 1 GAAGAUCACCUCAGAUACUGAUCGGCAGCAUUGUUGGCAAAACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCA ..........(((((...((((((((((...(((((......)))))....((....))..)))))).))))......((((......)))))))))..(((((((((...))))))))) ( -37.60) >DroSim_CAF1 12645 120 + 1 GAAGAUCACCUCAGAUACUGAUCGGCAGCAAUGUUGGCAAAACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCA ..........(((((...((((((((((...(((((......)))))....((....))..)))))).))))......((((......)))))))))..(((((((((...))))))))) ( -37.70) >DroEre_CAF1 12721 120 + 1 GAAGAUCACCUCAGAUACUGAUCGGCAGCAAUGUUGGCAAAACAACACCACCGAGUUCGAAUUGCCGCAUCAAAGUCGGCGACAAAUAUCGCUCUGACAUGCCCCGAUCCAAUCUGGGCA .........(((((((...((((((..(((((((.(((((..((((........))).)..)))))))))....((((((((......)))))..))).))).))))))..))))))).. ( -36.20) >DroYak_CAF1 12715 120 + 1 GAAGAUCACCUCAGAUACUGAUCGGCAGCAAUGUUGGCAAAACAACACCACCGAGUUCGAAUUGCCGCAUCAAAGCCUGCGACAAUAAUCGCUCUGACAUGCACCGAUCCAAUCUAGUCA ............((((...((((((..(((.(((((......))))).....((((....((((.((((........)))).))))....)))).....))).))))))..))))..... ( -29.80) >consensus GAAGAUCACCUCAGAUACUGAUCGGCAGCAAUGUUGGCAAAACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCUGGGCA .........(((((((...((((((.((((.(((((......))))).....((((.....(((((((......).))))))........)))).....))))))))))..))))))).. (-29.00 = -30.40 + 1.40)

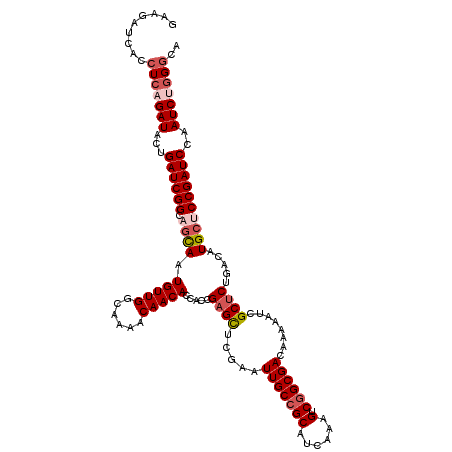

| Location | 4,100,724 – 4,100,844 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -44.40 |
| Consensus MFE | -39.00 |
| Energy contribution | -39.48 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4100724 120 - 27905053 UGCCCAGAUUGGAUCGGACCAUGUCAGAGCGAUUUUUGUCGCCGACUUUGAUGCGGCAAUUCGAGCGCGGUGGUGUUGUUUUGCCAACAUUACUGCCGAUCAGUAUCUGAGGUGAUCUUC .((((((((..((((((((...))).(..(((...(((((((..........))))))).)))..)((((((((((((......)))))))))))))))))...))))).)))....... ( -46.10) >DroSec_CAF1 12582 120 - 1 UGCCCCGAUUGGAUCGGAGCAUGUCAGAGCGAUUUUUGUCGCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAAUGCUGCCGAUCAGUAUCUGAGGUGAUCUUC .((.(((((...))))).))((.(((((.(((...(((((((..........))))))).))).(.((((..((((((((.....))))))))..)))).)....))))).))....... ( -43.10) >DroSim_CAF1 12645 120 - 1 UGCCCCGAUUGGAUCGGAGCAUGUCAGAGCGAUUUUUGUCGCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUC .((.(((((...))))).))((.((((((((((....))))).....(((((.(((((...((....))(..((((((......))))))..)))))))))))..))))).))....... ( -42.70) >DroEre_CAF1 12721 120 - 1 UGCCCAGAUUGGAUCGGGGCAUGUCAGAGCGAUAUUUGUCGCCGACUUUGAUGCGGCAAUUCGAACUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUC .((((((((..((((((((((((((((((((((((....((((((.(((((.........))))).)))))))))))))))))...))).)))).))))))...))))).)))....... ( -47.50) >DroYak_CAF1 12715 120 - 1 UGACUAGAUUGGAUCGGUGCAUGUCAGAGCGAUUAUUGUCGCAGGCUUUGAUGCGGCAAUUCGAACUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUC ....(((((..(((((((........((((((..(((((((((........))))))))))))..)))((..((((((......))))))..)))))))))...)))))........... ( -42.60) >consensus UGCCCAGAUUGGAUCGGAGCAUGUCAGAGCGAUUUUUGUCGCCGACUUUGAUGCGGCAAUUCGAGCUCGGUGGUGUUGUUUUGCCAACAUUGCUGCCGAUCAGUAUCUGAGGUGAUCUUC .((((((((..((((((.........(..(((...(((((((..........))))))).)))..).(((((((((((......)))))))))))))))))...))))).)))....... (-39.00 = -39.48 + 0.48)



| Location | 4,100,764 – 4,100,878 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4100764 114 + 27905053 AACAACACCACCGCGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGGUCCGAUCCAAUCUGGGCAGAUA------CGCUCAUAAGGAGCACGGAGCUGAUGUGCC ............(((((((..(((((((......).))))))........((((((.....(((((........))))).....------.((((.....)))).))))))))).)))). ( -35.00) >DroSec_CAF1 12622 114 + 1 AACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCAGAUA------CGCUCAUAAGGAGCACGGAGUGGAUGUGCC ..............((.((..(((((((......).))))))......((((((((...(((((((((...)))))))))....------.((((.....)))).)))))))).)).)). ( -39.30) >DroSim_CAF1 12685 114 + 1 AACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCAGAUA------CGCUCAUAAGGAGCACGGAGUGGAUGUGCC ..............((.((..(((((((......).))))))......((((((((...(((((((((...)))))))))....------.((((.....)))).)))))))).)).)). ( -39.30) >DroEre_CAF1 12761 114 + 1 AACAACACCACCGAGUUCGAAUUGCCGCAUCAAAGUCGGCGACAAAUAUCGCUCUGACAUGCCCCGAUCCAAUCUGGGCAGAUA------CGCUCAUAAGGAGCACGGAGCUGAUGUGCC ...........((....))..(((((((......).))))))...(((((((((((...(((((.(((...))).)))))....------.((((.....)))).)))))).)))))... ( -36.80) >DroYak_CAF1 12755 120 + 1 AACAACACCACCGAGUUCGAAUUGCCGCAUCAAAGCCUGCGACAAUAAUCGCUCUGACAUGCACCGAUCCAAUCUAGUCAGAUACUUGUCCGCUCAUAAGGAGCACGGAGCUGAUGUGCC .....(((....((((....((((.((((........)))).))))....))))...((.((.(((.....((((....))))........((((.....)))).))).))))..))).. ( -26.70) >consensus AACAACACCACCGAGCUCGAAUUGCCGCAUCAAAGUCGGCGACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCUGGGCAGAUA______CGCUCAUAAGGAGCACGGAGCUGAUGUGCC .....(((.............(((((((......).)))))).....(((((((((...(((((.(((...))).)))))...........((((.....)))).)))))).)))))).. (-27.90 = -28.54 + 0.64)



| Location | 4,100,804 – 4,100,918 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4100804 114 + 27905053 GACAAAAAUCGCUCUGACAUGGUCCGAUCCAAUCUGGGCAGAUA------CGCUCAUAAGGAGCACGGAGCUGAUGUGCCAUCCCUCAGCCGGAUGAUAACCAUCAGUAGGCAAUCUAUG (.((...(((((((((.....(((((........))))).....------.((((.....)))).)))))).))).)).)........(((.((((.....))))....)))........ ( -32.00) >DroSec_CAF1 12662 114 + 1 GACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCAGAUA------CGCUCAUAAGGAGCACGGAGUGGAUGUGCCAUCCCUCAGCCGGAUGAUAACCAUCAGUAGGCAAUCUAUG ..........((((((...(((((((((...)))))))))....------.((((.....)))).))))))(((((...)))))....(((.((((.....))))....)))........ ( -39.40) >DroSim_CAF1 12725 114 + 1 GACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCGGGGCAGAUA------CGCUCAUAAGGAGCACGGAGUGGAUGUGCCAUCCCUCAGCCGGAUGAUAACCAUCAGUAGGCAAUCUAUG ..........((((((...(((((((((...)))))))))....------.((((.....)))).))))))(((((...)))))....(((.((((.....))))....)))........ ( -39.40) >DroEre_CAF1 12801 114 + 1 GACAAAUAUCGCUCUGACAUGCCCCGAUCCAAUCUGGGCAGAUA------CGCUCAUAAGGAGCACGGAGCUGAUGUGCCAUCCCUCAGCCGGAUGAUAACCAUCAGUAGGCAAUCUAUG .....(((((((((((...(((((.(((...))).)))))....------.((((.....)))).)))))).)))))...........(((.((((.....))))....)))........ ( -37.70) >DroYak_CAF1 12795 120 + 1 GACAAUAAUCGCUCUGACAUGCACCGAUCCAAUCUAGUCAGAUACUUGUCCGCUCAUAAGGAGCACGGAGCUGAUGUGCCAUCCCUCAGCCGGAUGAUAAACAUCAGUAGGCAAUCUAUG ............((((((..................))))))...(((((.((((.....)))).....((((((((..(((((.......)))))....)))))))).)))))...... ( -31.37) >consensus GACAAAAAUCGCUCUGACAUGCUCCGAUCCAAUCUGGGCAGAUA______CGCUCAUAAGGAGCACGGAGCUGAUGUGCCAUCCCUCAGCCGGAUGAUAACCAUCAGUAGGCAAUCUAUG (.((...(((((((((...(((((.(((...))).)))))...........((((.....)))).)))))).))).)).)........(((.((((.....))))....)))........ (-29.76 = -30.20 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:02 2006