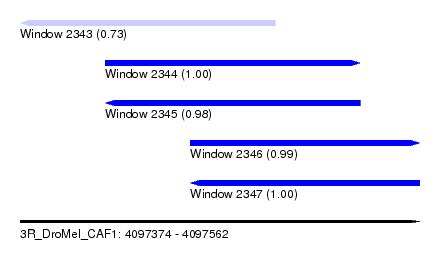
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,097,374 – 4,097,562 |
| Length | 188 |
| Max. P | 0.999849 |

| Location | 4,097,374 – 4,097,494 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4097374 120 - 27905053 UUCAACCGCAAAGGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGACUGGAAGGAUGAAUCUGGUCAGGUAGCUGAGGACAUUGCCUAAGAAGGGGAAAAUUCCCAGCAAAUUAGCAG ....(((.....((((((((.(((....))))))))))).......(((..((........))..))).))).((((.(((..((.(((.....))).))...))))))).......... ( -38.60) >DroSec_CAF1 9313 119 - 1 UACAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGGCUGAAACGAAGAAUCUGGUCAGGUAGCUGA-GACAUUGCCUAAGAAUGGGAAAAUUCCCAGCAAAUUAGCAG ....(((......(((((((.(((....))))))))))....((((..((.......))..))))....))).(((((-....((((.......(((((....))))))))).))))).. ( -32.91) >DroSim_CAF1 9344 120 - 1 UUCAACCGCAAAUGUCACUUAUAUUUAGCUGAAGUGACCACACAGAGGCUGAAACGAAAAAUCUGGUCAGGUAGCUGAGGACAUUGCCUAAGAAGGGGAAAAUUCCCAGCAAAUUAGCAG (((....((((.((((.((((..(((((((...(((....)))...))))))).......((((....))))...))))))))))))....))).((((....)))).((......)).. ( -28.70) >DroEre_CAF1 9360 118 - 1 UUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGCGGCUGGAAGGG--AAUCUGGUCAGGUAGCAAAGAACAUUGCCUAAGAGGGGGAAAAUUCCCAGCAAUUUAGCAG ((((.((((....(((((((.(((....))))))))))......)))).)))).(((--(((((..((((((((.........))))))..))..))....)))))).((......)).. ( -32.00) >DroYak_CAF1 9317 119 - 1 UUCAACCGCAAAUGUCACUUAAAGUUAGCUGAAGUGACCACACAGCGACUAGAAGGG-CAAUUUGGUCUGGUAGCGGGGGACAUUGCCGAAGGGAGGGAACAUUCCCAGCAAAUUAGCAG .((..((((....(((((((..((....)).)))))))........(((((((....-...))))))).....))))..)).....((....)).((((....)))).((......)).. ( -34.40) >consensus UUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGGCUGGAAGGA_GAAUCUGGUCAGGUAGCUGAGGACAUUGCCUAAGAAGGGGAAAAUUCCCAGCAAAUUAGCAG ....(((......(((((((.(((....))))))))))........(((((((........))))))).))).(((((.....((((........((((....)))).)))).))))).. (-25.44 = -25.84 + 0.40)

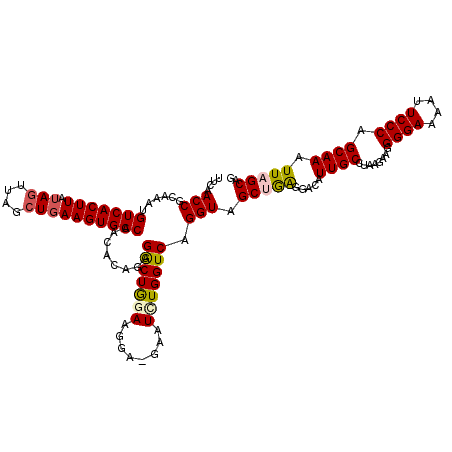
| Location | 4,097,414 – 4,097,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.99 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4097414 120 + 27905053 CCUCAGCUACCUGACCAGAUUCAUCCUUCCAGUCUCUGUGUGGUCACUUCAGCUAACUAUAAGUGACCUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCC ....((((...((((((.((.................)).))))))....))))..(((...(((((..(((((..(((((((....))))))).....)))))..)))))..))).... ( -29.23) >DroSec_CAF1 9353 119 + 1 C-UCAGCUACCUGACCAGAUUCUUCGUUUCAGCCUCUGUGUGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGUAAAUAUCAAUUUCCAUAAAAUGCAAUUGUCACCUUAGCGCC .-..((((...((((((.((.....((....))....)).))))))....))))..(((...((((((.(((((..((.((((....)))).)).....))))).))))))..))).... ( -27.60) >DroSim_CAF1 9384 120 + 1 CCUCAGCUACCUGACCAGAUUUUUCGUUUCAGCCUCUGUGUGGUCACUUCAGCUAAAUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUCCAUAAAAUGCAAUUGUCACCUUAGCGCC .....((....((((((.((.....((....))....)).)))))).....(((((......((((((.((((((((((.....)))))..........))))).)))))).))))))). ( -27.30) >DroEre_CAF1 9400 118 + 1 UCUUUGCUACCUGACCAGAUU--CCCUUCCAGCCGCUGUGUGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCC .....((....((((((.((.--..............)).)))))).....(((((......((((((.(((((..(((((((....))))))).....))))).)))))).))))))). ( -30.66) >DroYak_CAF1 9357 119 + 1 CCCCCGCUACCAGACCAAAUUG-CCCUUCUAGUCGCUGUGUGGUCACUUCAGCUAACUUUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCC ....(((((...(((((....(-(..........))....))))).................((((((.(((((..(((((((....))))))).....))))).))))))..))))).. ( -32.50) >consensus CCUCAGCUACCUGACCAGAUUC_UCCUUCCAGCCUCUGUGUGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCC .....((.....(((((.((.................)).)))))......(((((......((((((.(((((..(((((((....))))))).....))))).)))))).))))))). (-27.39 = -27.99 + 0.60)

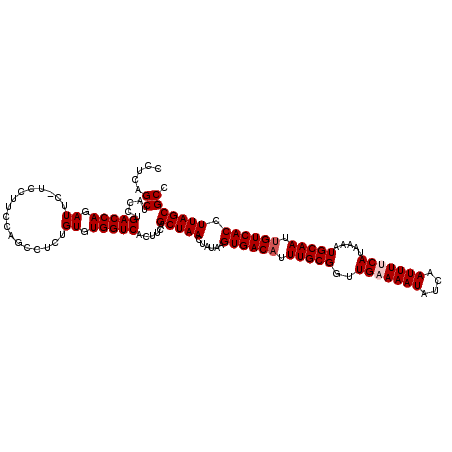
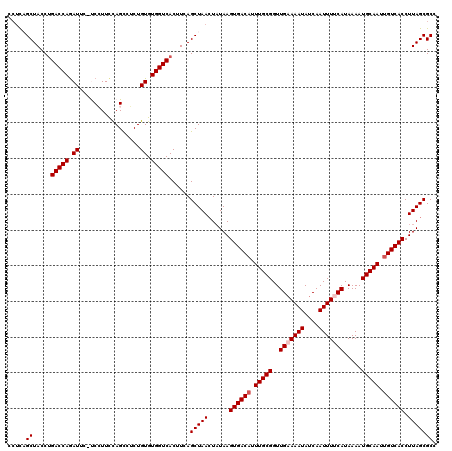
| Location | 4,097,414 – 4,097,534 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4097414 120 - 27905053 GGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAGGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGACUGGAAGGAUGAAUCUGGUCAGGUAGCUGAGG (((((...(....)....))......(((((((....)))))))(((.....((((((((.(((....))))))))))).......(((..((........))..))).))).))).... ( -35.30) >DroSec_CAF1 9353 119 - 1 GGCGCUAAGGUGACAAUUGCAUUUUAUGGAAAUUGAUAUUUACAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGGCUGAAACGAAGAAUCUGGUCAGGUAGCUGA-G ...((((((((((((.((((......((.((((....)))).))...)))).)))))))).))))((((((...((((((....((..((.......))..))))))))..)))))).-. ( -31.80) >DroSim_CAF1 9384 120 - 1 GGCGCUAAGGUGACAAUUGCAUUUUAUGGAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAUUUAGCUGAAGUGACCACACAGAGGCUGAAACGAAAAAUCUGGUCAGGUAGCUGAGG .......((((((((.((((......(((((((....)))))))...)))).))))))))...((((((((...((((((..(((...)))....((....))))))))..)))))))). ( -33.10) >DroEre_CAF1 9400 118 - 1 GGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGCGGCUGGAAGGG--AAUCUGGUCAGGUAGCAAAGA .((((((((((((((.((((......(((((((....)))))))...)))).)))))))).))))..((((..((....)).))))(((..((....--..))..))).....))..... ( -34.70) >DroYak_CAF1 9357 119 - 1 GGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAAAGUUAGCUGAAGUGACCACACAGCGACUAGAAGGG-CAAUUUGGUCUGGUAGCGGGGG ..(((((((((((((.((((......(((((((....)))))))...)))).)))))))).......((((..((....)).))))(((((((....-...)))))))...))))).... ( -34.50) >consensus GGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCACACAGAGGCUGGAAGGA_GAAUCUGGUCAGGUAGCUGAGG .......((((((((.((((......(((((((....)))))))...)))).))))))))....(((((((..(((....)))...(((((((........)))))))...))))))).. (-28.72 = -29.00 + 0.28)

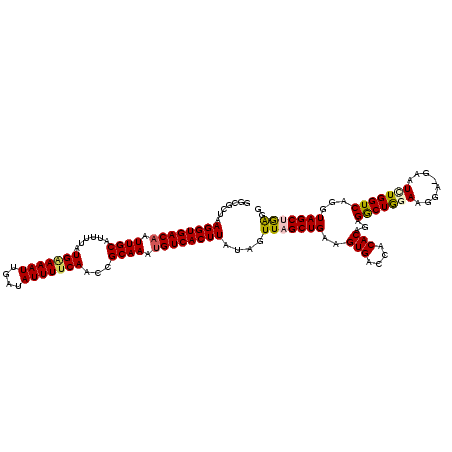
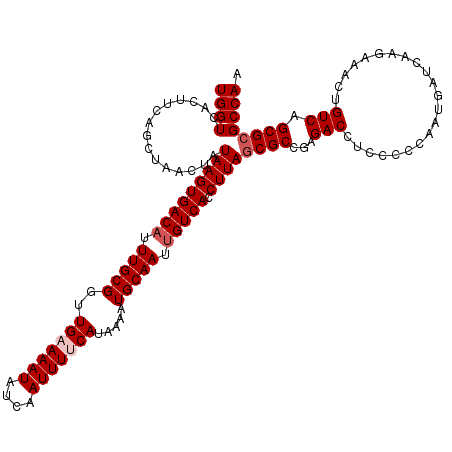
| Location | 4,097,454 – 4,097,562 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4097454 108 + 27905053 UGGUCACUUCAGCUAACUAUAAGUGACCUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAU------------GUCAGCGCGCCAA ((((...............((((((((..(((((..(((((((....))))))).....)))))..)))).))))((((...(((..........------------))).)))))))). ( -27.50) >DroSec_CAF1 9392 120 + 1 UGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGUAAAUAUCAAUUUCCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAUGAUCAAGAAACUGUCAGCGCGCCAA ((((...............(((((((((.(((((..((.((((....)))).)).....))))).))))).))))((((.((....))......(((.((......))))))))))))). ( -25.00) >DroSim_CAF1 9424 120 + 1 UGGUCACUUCAGCUAAAUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUCCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAUGAUCAAGAAACUGUCAGCGCGCCAA ((((...............(((((((((.((((((((((.....)))))..........))))).))))).))))((((.((....))......(((.((......))))))))))))). ( -25.10) >DroEre_CAF1 9438 120 + 1 UGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAUGAUCAAGAAACUGUCAGCGGGCCAA .((((.(....(((((......((((((.(((((..(((((((....))))))).....))))).)))))).)))))...).))))...(((..(((.((......)))))..))).... ( -33.20) >DroYak_CAF1 9396 120 + 1 UGGUCACUUCAGCUAACUUUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAUGAUCAAGAAACUGUCAGCGCGCCAA ((((..................((((((.(((((..(((((((....))))))).....))))).))))))....((((.((....))......(((.((......))))))))))))). ( -29.30) >consensus UGGUCACUUCAGCUAACUAUAAGUGACAUUUGCGGUUGAAAAUAUCAAUUUUCAUAAAAUGCAAUUGUCACCUUAGCGCCGAGACCUCCCCCAAUGAUCAAGAAACUGUCAGCGCGCCAA ((((...............(((((((((.(((((..(((((((....))))))).....))))).))))).))))((((...(((......................))).)))))))). (-24.77 = -25.57 + 0.80)

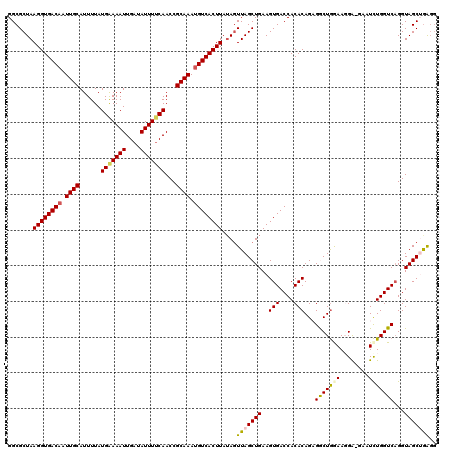
| Location | 4,097,454 – 4,097,562 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -32.72 |
| Energy contribution | -34.80 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4097454 108 - 27905053 UUGGCGCGCUGAC------------AUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAGGUCACUUAUAGUUAGCUGAAGUGACCA ...(((((((((.------------(((......))).)))))))(((((((.(....))))))))(((((((....)))))))...))...((((((((.(((....))))))))))). ( -35.50) >DroSec_CAF1 9392 120 - 1 UUGGCGCGCUGACAGUUUCUUGAUCAUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGGAAAUUGAUAUUUACAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCA (..(((((((((.(.((((((.(....).)))))).).)))))))..((((((((.((((......((.((((....)))).))...)))).)))))))).......))..)........ ( -35.10) >DroSim_CAF1 9424 120 - 1 UUGGCGCGCUGACAGUUUCUUGAUCAUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGGAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAUUUAGCUGAAGUGACCA (..(((((((((.(.((((((.(....).)))))).).)))))))..((((((((.((((......(((((((....)))))))...)))).)))))))).......))..)........ ( -39.00) >DroEre_CAF1 9438 120 - 1 UUGGCCCGCUGACAGUUUCUUGAUCAUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCA ....((((.((((((....))).))).))))...(((((((((((((((((((((.((((......(((((((....)))))))...)))).)))))))).))))..)))))...)))). ( -40.50) >DroYak_CAF1 9396 120 - 1 UUGGCGCGCUGACAGUUUCUUGAUCAUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAAAGUUAGCUGAAGUGACCA (..(((((((((.(.((((((.(....).)))))).).)))))))..((((((((.((((......(((((((....)))))))...)))).)))))))).......))..)........ ( -39.50) >consensus UUGGCGCGCUGACAGUUUCUUGAUCAUUGGGGGAGGUCUCGGCGCUAAGGUGACAAUUGCAUUUUAUGAAAAUUGAUAUUUUCAACCGCAAAUGUCACUUAUAGUUAGCUGAAGUGACCA (..(((((((((.(.((((((.(....).)))))).).)))))))..((((((((.((((......(((((((....)))))))...)))).)))))))).......))..)........ (-32.72 = -34.80 + 2.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:47 2006