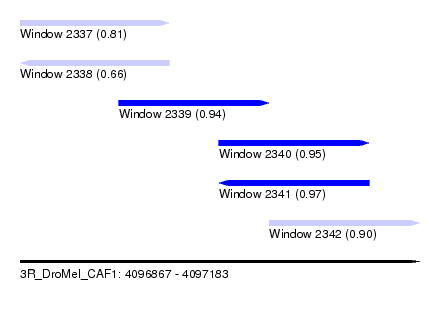
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,096,867 – 4,097,183 |
| Length | 316 |
| Max. P | 0.969377 |

| Location | 4,096,867 – 4,096,985 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4096867 118 + 27905053 CUCCGCUGAAUGGCUAUG--UAUGCGGACACGUGAUAGAACGGGGCCAGGGAUAUUAUCCUGCUUUCCCCUGGAGACGAAUCCAAGGCAAUUAACCCCUUUUGGCCCUCAUCUUCCACUC .(((((............--...)))))...(((..(((..((((((((((((...)))))......((.((((......)))).))..............)))))))..)))..))).. ( -35.56) >DroSec_CAF1 8826 118 + 1 CUCCGCUGAAUGGCUAUG--UAUGCGGACACGUGAUAGAACAGGGCCAGUGAUAUUAUCCUGCUUUCCCCUGGAGACGAAUCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUC .(((((............--...)))))...(((..(((..((((((((...........(((((.....((((......)))))))))...........))))))))..)))..))).. ( -35.51) >DroSim_CAF1 8857 118 + 1 CUCCGCUGAAUGGCUAUG--UAUGCGGACACGUGAUAGAACAGGGCCAGGCAUAUUAUCCUGCUUUCCCCUGGAGACGAAUCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUC .(((((............--...)))))...(((..(((..(((((((((((........)))....((.((((......)))).)).............))))))))..)))..))).. ( -36.76) >DroEre_CAF1 8858 118 + 1 CUCCGCUGAAUAGCUAUG--UAUACGGACACGUGAUAGAACAGGGUCAGGGAAAUUAUCCUGCUUUCCCCUGGAGACGAAGCCUAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUC .((((.((.(((....))--).))))))...(((..(((..(((((((((((.....)))(((((.....((....))......)))))...........))))))))..)))..))).. ( -31.00) >DroYak_CAF1 8802 120 + 1 CUCCGCUGAAUAGCUAUGUGUGUACGGACACGUGAUAGAACAGGGCAAGGGAAAUUAUCCUGCUUUCCCCUGGAGACGAAACAGAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUC ....(((....)))...((((......))))(((..(((..(((((..((((((.........)))))).((....))...((((((..........)))))))))))..)))..))).. ( -32.40) >consensus CUCCGCUGAAUGGCUAUG__UAUGCGGACACGUGAUAGAACAGGGCCAGGGAUAUUAUCCUGCUUUCCCCUGGAGACGAAUCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUC .(((((.................)))))...(((..(((..((((((((((.........((((((....((....)).....)))))).........))))))))))..)))..))).. (-29.94 = -30.54 + 0.60)

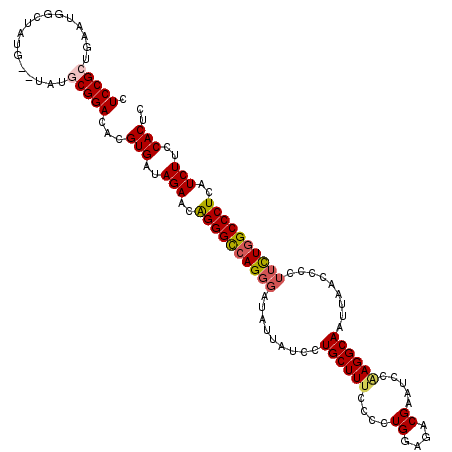
| Location | 4,096,867 – 4,096,985 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -33.37 |
| Energy contribution | -34.93 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4096867 118 - 27905053 GAGUGGAAGAUGAGGGCCAAAAGGGGUUAAUUGCCUUGGAUUCGUCUCCAGGGGAAAGCAGGAUAAUAUCCCUGGCCCCGUUCUAUCACGUGUCCGCAUA--CAUAGCCAUUCAGCGGAG ((((((...(((.(((((....(((((...((.(((((((......))))))).))..((((........)))))))))((......))).)))).))).--.....))))))....... ( -41.30) >DroSec_CAF1 8826 118 - 1 GAGUGGAAGAUGAGGGCCAGAAGGGGUUAAUUGCCUUGGAUUCGUCUCCAGGGGAAAGCAGGAUAAUAUCACUGGCCCUGUUCUAUCACGUGUCCGCAUA--CAUAGCCAUUCAGCGGAG ..((((.(((..((((((((.....(((..((.(((((((......))))))).)))))..(((...))).))))))))..))).))))...(((((...--............))))). ( -42.26) >DroSim_CAF1 8857 118 - 1 GAGUGGAAGAUGAGGGCCAGAAGGGGUUAAUUGCCUUGGAUUCGUCUCCAGGGGAAAGCAGGAUAAUAUGCCUGGCCCUGUUCUAUCACGUGUCCGCAUA--CAUAGCCAUUCAGCGGAG ..((((.(((..((((((((..........((.(((((((......))))))).)).(((........)))))))))))..))).))))...(((((...--............))))). ( -44.66) >DroEre_CAF1 8858 118 - 1 GAGUGGAAGAUGAGGGCCAGAAGGGGUUAAUUGCCUAGGCUUCGUCUCCAGGGGAAAGCAGGAUAAUUUCCCUGACCCUGUUCUAUCACGUGUCCGUAUA--CAUAGCUAUUCAGCGGAG ..((((.((((((((.((.....((((.....)))).)))))))))).(((((.....((((........)))).))))).....))))...(((((...--............))))). ( -36.36) >DroYak_CAF1 8802 120 - 1 GAGUGGAAGAUGAGGGCCAGAAGGGGUUAAUUGCCUCUGUUUCGUCUCCAGGGGAAAGCAGGAUAAUUUCCCUUGCCCUGUUCUAUCACGUGUCCGUACACACAUAGCUAUUCAGCGGAG ..((((.(((..(((((....((((((.....))))))...........((((((((.........)))))))))))))..))).))))((((......))))...(((....))).... ( -38.00) >consensus GAGUGGAAGAUGAGGGCCAGAAGGGGUUAAUUGCCUUGGAUUCGUCUCCAGGGGAAAGCAGGAUAAUAUCCCUGGCCCUGUUCUAUCACGUGUCCGCAUA__CAUAGCCAUUCAGCGGAG ..((((.(((..((((((((.....(((..((.((((((........)))))).))))).(((.....)))))))))))..))).))))...(((((.................))))). (-33.37 = -34.93 + 1.56)

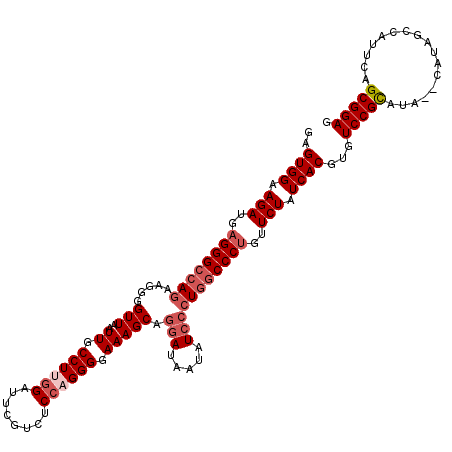
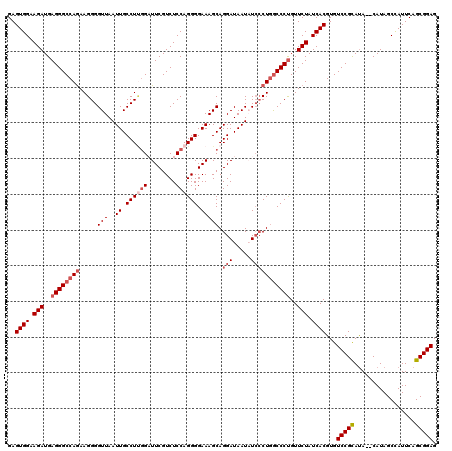
| Location | 4,096,945 – 4,097,064 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -28.86 |
| Energy contribution | -29.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4096945 119 + 27905053 UCCAAGGCAAUUAACCCCUUUUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUU-UCGGGUCGAUUUAUCAAAUAAUUCCACGAAUACCUAGAAUUCGAAAUAGA .....((.(((((..........((((((...............))))))(.((((((.(((.((....-.)).))))))))).)...))))))).(((((.......)))))....... ( -27.06) >DroSec_CAF1 8904 119 + 1 UCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUU-UCGAGUCGAUUUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGA .....((.(((((..........((((((...............))))))(.((((((.(((.((....-.)).))))))))).)...))))))).(((((((...)))))))....... ( -30.06) >DroSim_CAF1 8935 119 + 1 UCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUU-UCGAGUCGAUUUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGA .....((.(((((..........((((((...............))))))(.((((((.(((.((....-.)).))))))))).)...))))))).(((((((...)))))))....... ( -30.06) >DroEre_CAF1 8936 119 + 1 GCCUAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUU-UCGGGUCGAUUUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGA ..(((((.(((((..........((((((...............))))))(.((((((.(((.((....-.)).))))))))).)...))))))).(((((((...)))))))...))). ( -30.86) >DroYak_CAF1 8882 120 + 1 ACAGAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUUUUCGGGUCGAUUUAUCAAAUAAUUCCACGAAUUCCUGGAACUCGAAAUAGA .((((((..........))))))((((((...............))))))(.((((((.(((.((......)).))))))))).)......(((((.(....).)))))........... ( -29.96) >consensus UCCAAGGCAAUUAACCCCUUCUGGCCCUCAUCUUCCACUCCUGCGAGGGCGUUAAGUCAGACACGAGUU_UCGGGUCGAUUUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGA .....((.(((((..........((((((...............))))))(.((((((.(((.(((....))).))))))))).)...))))))).(((((((...)))))))....... (-28.86 = -29.26 + 0.40)

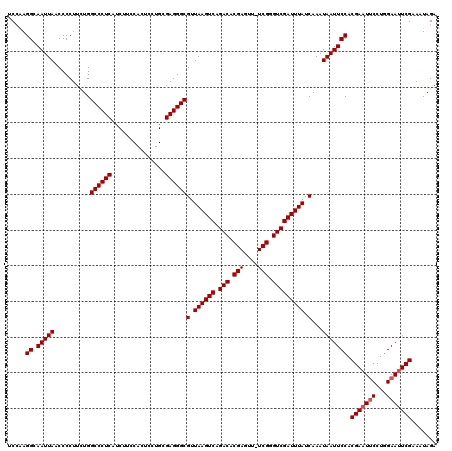
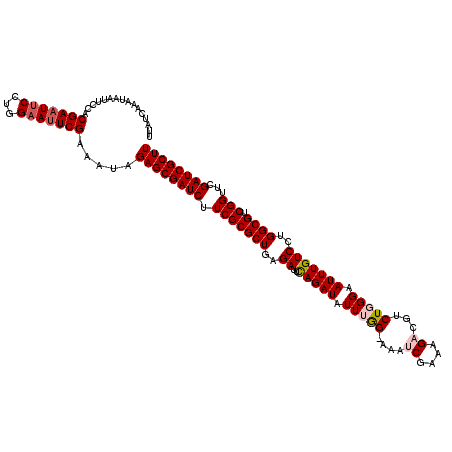
| Location | 4,097,024 – 4,097,143 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -29.58 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
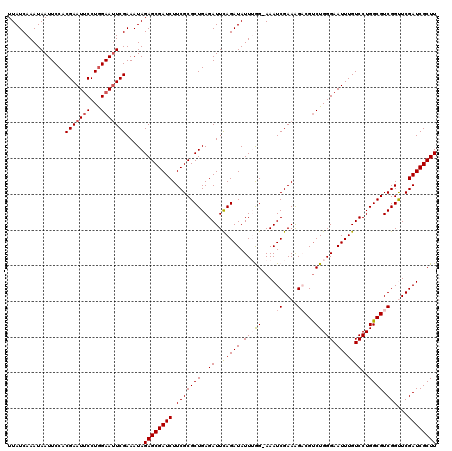
>3R_DroMel_CAF1 4097024 119 + 27905053 UUAUCAAAUAAUUCCACGAAUACCUAGAAUUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUCAGAUAUUUGG-AAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUU ................(((((.......))))).....((((((((.(((((((..((..(((((.((..(-(..((....))..))..)).)))))))..)))).)))...)))))))) ( -35.80) >DroSec_CAF1 8983 120 + 1 UUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUUAGAUAUUAGGGAAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGCUCGAUCGCUU .((((...........(((((((...)))))))...(((.(((.....))).))).......))))...(((..(((((..((((.(..(((.....)))..)))))...)))))..))) ( -34.20) >DroSim_CAF1 9014 120 + 1 UUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUUAGAUAUUAGGAAAAUCGAAGGACGUCUGGGAAUUUGUCCUGGCGUCGGCUCGAUCGCUU ................(((((((...))))))).....((((((((...((.((..(((((...........)))))..))((((.(..(((.....)))..))))).))..)))))))) ( -34.80) >DroEre_CAF1 9015 119 + 1 UUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUCAGAUAUUUAG-AAAUCGAAAGCCGCCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUU ................(((((((...))))))).....((((((((......(((.....))).......(-((.((((....((((.((((.....))))))))))))))))))))))) ( -34.00) >DroYak_CAF1 8962 119 + 1 UUAUCAAAUAAUUCCACGAAUUCCUGGAACUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUCAGAUAUUUAG-AAAUCGAAAGCCGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUU ...........(((((.(....).))))).........((((((((.(((((((..((..(((((.(((((-(...(....)...)))))).)))))))..)))).)))...)))))))) ( -34.60) >consensus UUAUCAAAUAAUUCCACGAAUUCCUGGAAUUCGAAAUAGAGCGAUCUUCGCGCUGAGAUUCAGAUAUUUGG_AAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUU ................(((((((...))))))).....((((((((.(((((((..((..(((((.(((((....((....))...))))).)))))))..)))).)))...)))))))) (-29.58 = -30.30 + 0.72)

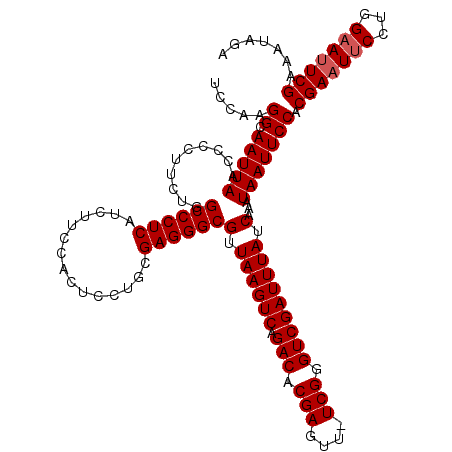
| Location | 4,097,024 – 4,097,143 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
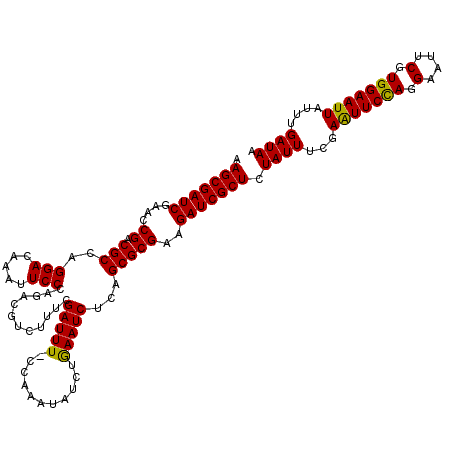
>3R_DroMel_CAF1 4097024 119 - 27905053 AAGCGAUCGAACCGACGCCAGGACAAAUUCCCAGACGUCUUUCGAUUU-CCAAAUAUCUGAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAAUUCUAGGUAUUCGUGGAAUUAUUUGAUAA ....(((((((..(((((..(((.....)))..).)))).)))))))(-((((((((((((((.((((.(((.....))).)).....)))))).)))))))..))))............ ( -28.10) >DroSec_CAF1 8983 120 - 1 AAGCGAUCGAGCCGACGCCAGGACAAAUUCCCAGACGUCUUUCGAUUUCCCUAAUAUCUAAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAAUUCCAGGAAUUCGUGGAAUUAUUUGAUAA .(((((((..((((((((..(((.....)))..).))))....(((((...........)))))...).))...))))))).((((...(((((((.(....).)))))))....)))). ( -31.30) >DroSim_CAF1 9014 120 - 1 AAGCGAUCGAGCCGACGCCAGGACAAAUUCCCAGACGUCCUUCGAUUUUCCUAAUAUCUAAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAAUUCCAGGAAUUCGUGGAAUUAUUUGAUAA .(((((((..((((((((..(((.....)))..).))))....(((((...........)))))...).))...))))))).((((...(((((((.(....).)))))))....)))). ( -31.70) >DroEre_CAF1 9015 119 - 1 AAGCGAUCGAACCGACGCCAGGACAAAUUCCCAGGCGGCUUUCGAUUU-CUAAAUAUCUGAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAAUUCCAGGAAUUCGUGGAAUUAUUUGAUAA .((.(((((((....((((.(((.....)))..))))...))))))).-))...((((((((....((.(((.....))).))..))))(((((((.(....).)))))))....)))). ( -33.60) >DroYak_CAF1 8962 119 - 1 AAGCGAUCGAACCGACGCCAGGACAAAUUCCCAGACGGCUUUCGAUUU-CUAAAUAUCUGAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAGUUCCAGGAAUUCGUGGAAUUAUUUGAUAA .(((((((....((.(((..(((.....))).....((..((((((..-......))).)))..)).)))))..))))))).((((...(((((((.(....).)))))))....)))). ( -30.00) >consensus AAGCGAUCGAACCGACGCCAGGACAAAUUCCCAGACGUCUUUCGAUUU_CCAAAUAUCUGAAUCUCAGCGCGAAGAUCGCUCUAUUUCGAAUUCCAGGAAUUCGUGGAAUUAUUUGAUAA .(((((((....((.(((..(((.....)))............(((((...........)))))...)))))..))))))).((((...(((((((.(....).)))))))....)))). (-27.88 = -27.32 + -0.56)

| Location | 4,097,064 – 4,097,183 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.44 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
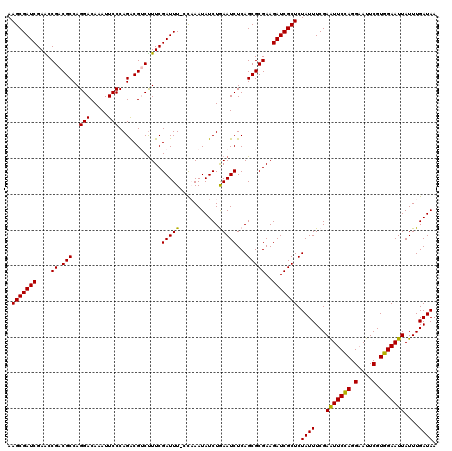
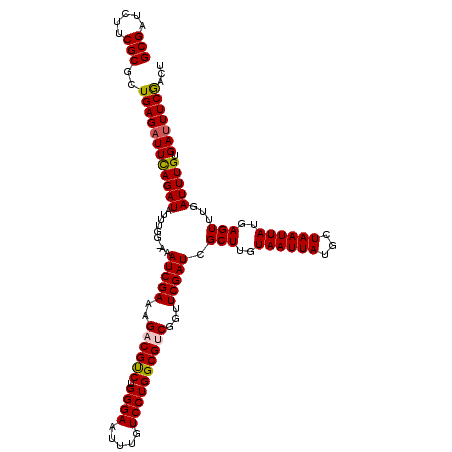
>3R_DroMel_CAF1 4097064 119 + 27905053 GCGAUCUUCGCGCUGAGAUUCAGAUAUUUGG-AAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGAUUUCGACU (((.....)))..((((((((((((......-..((((((.((((.(..(((.....)))..)))))..)))))).(((..((((((...))))))..)))...))))).)))))))... ( -36.70) >DroSec_CAF1 9023 120 + 1 GCGAUCUUCGCGCUGAGAUUUAGAUAUUAGGGAAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGCUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGAUUUCGACU (((.....)))..(((((((.....(((((....(((((..((((.(..(((.....)))..)))))...))))).(((..((((((...))))))..))))))))....)))))))... ( -33.30) >DroSim_CAF1 9054 120 + 1 GCGAUCUUCGCGCUGAGAUUUAGAUAUUAGGAAAAUCGAAGGACGUCUGGGAAUUUGUCCUGGCGUCGGCUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGAUUUCGACU (((.....)))..(((((((.....(((((....(((((..((((.(..(((.....)))..)))))...))))).(((..((((((...))))))..))))))))....)))))))... ( -32.20) >DroEre_CAF1 9055 119 + 1 GCGAUCUUCGCGCUGAGAUUCAGAUAUUUAG-AAAUCGAAAGCCGCCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGACUUCAACU (((.....))).(((.....)))....(((.-(((((((.(((((((..(((.....)))..).).))))).....(((..((((((...))))))..)))))))))).)))........ ( -31.50) >DroYak_CAF1 9002 119 + 1 GCGAUCUUCGCGCUGAGAUUCAGAUAUUUAG-AAAUCGAAAGCCGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGAUUUCAACU (((.....)))..((((((((((((......-..(((((..((((.(..(((.....)))..)...))))))))).(((..((((((...))))))..)))...))))).)))))))... ( -33.50) >consensus GCGAUCUUCGCGCUGAGAUUCAGAUAUUUGG_AAAUCGAAAGACGUCUGGGAAUUUGUCCUGGCGUCGGUUCGAUCGCUUGUAAUUAUGCUAAUUAUGAGUUUGAUUUGUGAUUUCGACU (((.....)))..((((((((((((.........(((((..((((((.((((.....))))))))))...))))).(((..((((((...))))))..)))...))))).)))))))... (-32.48 = -32.44 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:43 2006