
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,095,182 – 4,095,462 |
| Length | 280 |
| Max. P | 0.979126 |

| Location | 4,095,182 – 4,095,302 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -34.80 |
| Energy contribution | -34.68 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095182 120 - 27905053 UCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGUCCUAGCCCAGUUUUUGGAACACGAAGACUUGGAGCACCUGAAUCAACAGUUGAUUGUAGGCGGCAACAGCAUUGAAGAGG .(((....((((((((..(((....)))..)).))).)))....((((((((((((.....))))))).))).((.(((((((((.....))))).))))..))....))......))). ( -35.40) >DroSec_CAF1 7100 120 - 1 UCUCACCAACCCGGGAACUGGCGCGCCAAAUUUCCGAGGUCCUAGCCCAGUUUCUGGAACACGAAGACUUGGAGCACCUGAAUCAACAGUUGAUUGUAGGUGGCAACAGCAUAGAAGAGG .(((.((((.(((((((((((.(((((..........)))....)))))))))))))..(.....)..))))..(((((((((((.....))))).))))))((....))......))). ( -36.80) >DroSim_CAF1 7137 120 - 1 UCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGUCCUAGCCCAGUUUCUGGAACACGAAGACUUGGAGCACCUGAAUCAACAGUUGAUUGUAGGUGGCAACAGCAUAGAAGAGG .(((.((((.(((((((((((.(((((..........)))....)))))))))))))..(.....)..))))..(((((((((((.....))))).))))))((....))......))). ( -36.80) >DroEre_CAF1 7144 120 - 1 UCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGUUCUAGCCCAGUUUCUGGAGCACGAAGACCUCGAGCACCUAAAUCAACAGUUGAUUGUAGGUGGCAACAGCAUAGAAGAGG .(((......(((((((((((.((....((((.....))))...))))))))))))).((.(((.....)))..(((((((((((.....))))).))))))......))......))). ( -40.00) >DroYak_CAF1 7101 120 - 1 UCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGUUUUAGCCCAGUUUCUGGAGCACGAAGACCUCGAGCACCUGAAUCAACAGUUGAUUGUAGGUGGCAACAGCAUAGAAGAGG .(((......(((((((((((.((...(((((.....)))))..))))))))))))).((.(((.....)))..(((((((((((.....))))).))))))......))......))). ( -41.40) >consensus UCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGUCCUAGCCCAGUUUCUGGAACACGAAGACUUGGAGCACCUGAAUCAACAGUUGAUUGUAGGUGGCAACAGCAUAGAAGAGG .(((......(((((((((((.(((((..........)))....)))))))))))))................((((((((((((.....))))).)))))(....).))......))). (-34.80 = -34.68 + -0.12)

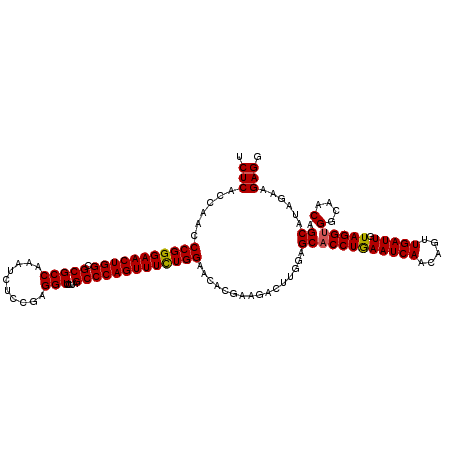

| Location | 4,095,222 – 4,095,342 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -51.32 |
| Consensus MFE | -44.50 |
| Energy contribution | -44.86 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095222 120 + 27905053 CAGGUGCUCCAAGUCUUCGUGUUCCAAAAACUGGGCUAGGACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGACGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUC ..(((((.(((((((((((.(.(((.............))).).))))))))))).)))))((..((((.((((......))))(.((((.((((......))))))))).))))..)). ( -52.32) >DroSec_CAF1 7140 120 + 1 CAGGUGCUCCAAGUCUUCGUGUUCCAGAAACUGGGCUAGGACCUCGGAAAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUAAGCGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUC ..(((((.((((((.((((.(.(((((........)).))).).)))).)))))).)))))((..((((((...((..........))(((((.........))))).)).))))..)). ( -44.40) >DroSim_CAF1 7177 120 + 1 CAGGUGCUCCAAGUCUUCGUGUUCCAGAAACUGGGCUAGGACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUC ..(((((.(((((((((((.(.(((((........)).))).).))))))))))).)))))((..((((.(((.....)))..((.(((((((.........)))))))))))))..)). ( -56.10) >DroEre_CAF1 7184 120 + 1 UAGGUGCUCGAGGUCUUCGUGCUCCAGAAACUGGGCUAGAACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUCAGGGCACCUAUUUCCUUGGGGCCCCAGGGGGUCUC (((((((((..((((.((..((.((((...))))))...((((.(((.((.((((....)))).))))))))).....)).)))).))))))))).......(((((((....))))))) ( -53.60) >DroYak_CAF1 7141 120 + 1 CAGGUGCUCGAGGUCUUCGUGCUCCAGAAACUGGGCUAAAACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCACCUAUUUCCUUGGGGCCCCAUGGGGUCUC .((((((((...(((.((..((.((((...))))))...((((.(((.((.((((....)))).))))))))).....)).)))..))))))))........(((((((....))))))) ( -50.20) >consensus CAGGUGCUCCAAGUCUUCGUGUUCCAGAAACUGGGCUAGGACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUC .((((((((...(((.((..((.((((...))))))...((((.(((.((.((((....)))).))))))))).....)).)))..))))))))........(((((((....))))))) (-44.50 = -44.86 + 0.36)



| Location | 4,095,262 – 4,095,382 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -50.62 |
| Consensus MFE | -41.36 |
| Energy contribution | -42.08 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095262 120 + 27905053 ACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGACGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUCUGGAGGAUUUCUAGCAUGGGCACAAGG (((.(((.((.((((....)))).))))))))....(((((..((.((((.((((......))))))))..))..)))))((((((((....((((((...))))))....)))))))). ( -52.20) >DroSec_CAF1 7180 120 + 1 ACCUCGGAAAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUAAGCGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUUUGCAGGAUUUCUAGCAUGGGCACAAGG .((..((((((..(((((..((.(((((.....)).))).))....)))))...))))))..))(((((....))))).(((((((((.....(((.((....)).)))..))))))))) ( -51.10) >DroSim_CAF1 7217 120 + 1 ACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUUUGCAGGAUUUCUAGCAUCGGCACAAGG (((.(((.((.((((....)))).))))))))....(((((..((.(((((((.........)))))))..))..)))))(((((((((....(((.((....)).))).))))))))). ( -56.80) >DroEre_CAF1 7224 120 + 1 ACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUCAGGGCACCUAUUUCCUUGGGGCCCCAGGGGGUCUCCUUGUGGCGUCUCUGCAGGAUUUCUAGCAUGGGCACAAGG (((.(((.((.((((....)))).))))))))....(((((..((.((((.((((......))))))))..))..)))))(((((...((((.(((.((....)).))).))))))))). ( -45.10) >DroYak_CAF1 7181 120 + 1 ACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCACCUAUUUCCUUGGGGCCCCAUGGGGUCUCCUUGUGGCGCCUCUGCAGGAUUUCUAGCAUGGGCACAAGG (((.(((.((.((((....)))).))))))))....(((((.(((.((((.((((......)))))))))))...)))))(((((...((((.(((.((....)).))).))))))))). ( -47.90) >consensus ACCUCGGAGAUUUGGCGCGCCAGUUCCCGGGUUGGUGAGAUGAUGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUCUGCAGGAUUUCUAGCAUGGGCACAAGG (((.(((.((.((((....)))).))))))))....(((((..((.((((.((((......))))))))..))..)))))((((((((.....(((.((....)).)))..)))))))). (-41.36 = -42.08 + 0.72)



| Location | 4,095,262 – 4,095,382 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -44.04 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.36 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095262 120 - 27905053 CCUUGUGCCCAUGCUAGAAAUCCUCCAGAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGUGCCCUCGUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGU .((((((((....((.((.....)).))....))))))))((((...((.(((((.((.........)).)))))))...))))....((((((((..(((....)))..)).))).))) ( -42.80) >DroSec_CAF1 7180 120 - 1 CCUUGUGCCCAUGCUAGAAAUCCUGCAAAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGGGCGCUUAUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUUUCCGAGGU (((((((((..(((.((.....))))).....)))))))))...(((....)))..((..((((((.((.(((((.....((((........))))...))))).))..))))))..)). ( -46.20) >DroSim_CAF1 7217 120 - 1 CCUUGUGCCGAUGCUAGAAAUCCUGCAAAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGGGCCCUCAUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGU .(((((((((.(((.((.....)))))....)))))))))((((....((((((((((.........))))))).)))..))))....((((((((..(((....)))..)).))).))) ( -52.10) >DroEre_CAF1 7224 120 - 1 CCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGCCACAAGGAGACCCCCUGGGGCCCCAAGGAAAUAGGUGCCCUGAUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGU (((((((.(..(((.((.....)))))..).))).....(((((..((.(((....))).)).....(((((((......((((........))))...)).)))))...))))))))). ( -36.00) >DroYak_CAF1 7181 120 - 1 CCUUGUGCCCAUGCUAGAAAUCCUGCAGAGGCGCCACAAGGAGACCCCAUGGGGCCCCAAGGAAAUAGGUGCCCUCAUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGU (((((((((..(((.((.....)))))..))))).....(((((..((.(((....))).)).....(((((((......((((........))))...)).)))))...))))))))). ( -43.10) >consensus CCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGUGCCCUCAUCAUCUCACCAACCCGGGAACUGGCGCGCCAAAUCUCCGAGGU .((((((((..(((.((.....))))).....))))))))((((......(((((.((.........)).))))).....))))....((((((((..(((....)))..)).))).))) (-35.72 = -36.36 + 0.64)



| Location | 4,095,302 – 4,095,422 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -36.52 |
| Energy contribution | -37.72 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095302 120 + 27905053 UGACGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUCUGGAGGAUUUCUAGCAUGGGCACAAGGAAGGCUAGCGUUUUACCACUACCAGUAACAGCCUCCGCUG ..(((.((((.((((......)))))))))(((.((((((((((((((....((((((...))))))....))))))))).)))))..))).............))..((((....)))) ( -45.40) >DroSec_CAF1 7220 120 + 1 UGAUAAGCGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUUUGCAGGAUUUCUAGCAUGGGCACAAGGAAGGCUAGUGUUUUACCACUACCAGUAACAGCCUCCGCCG ......(((((((.........))))).....(((((.((((((((((.....(((.((....)).)))..)))))))))).((.(((((......))))))).......))).)))).. ( -46.10) >DroSim_CAF1 7257 120 + 1 UGAUGAGGGCCCCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUUUGCAGGAUUUCUAGCAUCGGCACAAGGAAGGUUAGUGUUUUACCACUACCAGUAACUGCCUCCGCUG ...((.(((((((.........)))))))))((((((.(((((((((((....(((.((....)).))).))))))))))).(((.((((......))))))).......))).)))... ( -56.40) >DroEre_CAF1 7264 120 + 1 UGAUCAGGGCACCUAUUUCCUUGGGGCCCCAGGGGGUCUCCUUGUGGCGUCUCUGCAGGAUUUCUAGCAUGGGCACAAGGAAAGCCAGUGUUUUACCACUACCAGUAACCGCCUCCGCGG ......((((.((((......))))))))..((((.(.(((((((...((((.(((.((....)).))).))))))))))).).))((((......)))).)).....((((....)))) ( -43.40) >DroYak_CAF1 7221 120 + 1 UGAUGAGGGCACCUAUUUCCUUGGGGCCCCAUGGGGUCUCCUUGUGGCGCCUCUGCAGGAUUUCUAGCAUGGGCACAAGGAAAGCCAGUGUUUUACCACUACCAGUAACUGCCUCCGCGG ..(((.((((.((((......)))))))))))(((((.(((((((...((((.(((.((....)).))).)))))))))))..)))((((......)))).)).....((((....)))) ( -45.40) >consensus UGAUGAGGGCACCUAUUUCCUUGGGGCCCCACGGGGUCUCCUUGUGCCGUCUCUGCAGGAUUUCUAGCAUGGGCACAAGGAAGGCUAGUGUUUUACCACUACCAGUAACAGCCUCCGCGG ......((((.((((......))))))))...((((((((((((((((.....(((.((....)).)))..))))))))).)))))((((......)))).)).....((((....)))) (-36.52 = -37.72 + 1.20)



| Location | 4,095,302 – 4,095,422 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -46.40 |
| Consensus MFE | -37.54 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095302 120 - 27905053 CAGCGGAGGCUGUUACUGGUAGUGGUAAAACGCUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUCCAGAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGUGCCCUCGUCA ..((((.((........(((((((......)))).)))(((((((((((....((.((.....)).))....))))))))))).))))))(((((.((.........)).)))))..... ( -44.70) >DroSec_CAF1 7220 120 - 1 CGGCGGAGGCUGUUACUGGUAGUGGUAAAACACUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUGCAAAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGGGCGCUUAUCA ..((((.((........(((((((......)))).)))(((((((((((..(((.((.....))))).....))))))))))).))))))((.(((((.........))))).))..... ( -47.70) >DroSim_CAF1 7257 120 - 1 CAGCGGAGGCAGUUACUGGUAGUGGUAAAACACUAACCUUCCUUGUGCCGAUGCUAGAAAUCCUGCAAAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGGGCCCUCAUCA ..((((.(((.......(((((((......)))).))).(((((((((((.(((.((.....)))))....)))))))))))).))))))((((((((.........))))))))..... ( -55.40) >DroEre_CAF1 7264 120 - 1 CCGCGGAGGCGGUUACUGGUAGUGGUAAAACACUGGCUUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGCCACAAGGAGACCCCCUGGGGCCCCAAGGAAAUAGGUGCCCUGAUCA ...(((.((.((((...(.(((((......))))).)..(((((((((...(((.((.....))))).....).)))))))))))))))))((((.((.........)).))))...... ( -39.80) >DroYak_CAF1 7221 120 - 1 CCGCGGAGGCAGUUACUGGUAGUGGUAAAACACUGGCUUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGGCGCCACAAGGAGACCCCAUGGGGCCCCAAGGAAAUAGGUGCCCUCAUCA ....(..((((.(((..(.(((((......))))).)((((((((((((..(((.((.....)))))..))))).....((.(.(((....)))).)))))))))))).))))..).... ( -44.40) >consensus CAGCGGAGGCAGUUACUGGUAGUGGUAAAACACUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGGCACAAGGAGACCCCGUGGGGCCCCAAGGAAAUAGGUGCCCUCAUCA ..((((.((........(((((((......))))).))(((((((((((..(((.((.....))))).....))))))))))).))))))(((((.((.........)).)))))..... (-37.54 = -38.10 + 0.56)



| Location | 4,095,342 – 4,095,462 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -45.22 |
| Consensus MFE | -35.54 |
| Energy contribution | -36.42 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4095342 120 - 27905053 ACGGCGGCCAUACCGCUACUUCUGGCCCGGAAGGAUGUCUCAGCGGAGGCUGUUACUGGUAGUGGUAAAACGCUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUCCAGAGACGGCACAAG ..((((((..((((((((((..((((((....))..(((((....))))).))))..))))))))))....))).)))...((((((((....((.((.....)).))....)))))))) ( -47.90) >DroSec_CAF1 7260 120 - 1 ACGGCGGCCAUUCCGCUGCUCCUGGCCCGGAAAGAUGUCUCGGCGGAGGCUGUUACUGGUAGUGGUAAAACACUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUGCAAAGACGGCACAAG ..(((((((..(((((((.(((......))).((....))))))))))))))))...(((((((......)))).)))...((((((((..(((.((.....))))).....)))))))) ( -43.30) >DroSim_CAF1 7297 120 - 1 ACGGCGGCCAUUCCGCUGCUCCUGGCCCGGAAAGAUGUCUCAGCGGAGGCAGUUACUGGUAGUGGUAAAACACUAACCUUCCUUGUGCCGAUGCUAGAAAUCCUGCAAAGACGGCACAAG ..(((.(((..(((((((.(((......))).((....)))))))))))).)))...(((((((......)))).)))...(((((((((.(((.((.....)))))....))))))))) ( -42.90) >DroEre_CAF1 7304 120 - 1 ACGGCGGCCAUUCCGCUGCUCCUGGCCCGAAAGGAUGUCUCCGCGGAGGCGGUUACUGGUAGUGGUAAAACACUGGCUUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGCCACAAG .((((((.....))))))((..((((((....))..(((((.((((....((((.(((((((((......))).(((.........)))..)))))).)))))))).)))))))))..)) ( -45.20) >DroYak_CAF1 7261 120 - 1 ACGGCGGCCAUUCCGCUGCUCCUGGCCCGGAAGGAUGUCUCCGCGGAGGCAGUUACUGGUAGUGGUAAAACACUGGCUUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGGCGCCACAAG ..((((((((((((((((((..((((((....)).((((((....))))))))))..)))))))).)).....)))))........(((..(((.((.....)))))..))))))..... ( -46.80) >consensus ACGGCGGCCAUUCCGCUGCUCCUGGCCCGGAAGGAUGUCUCAGCGGAGGCAGUUACUGGUAGUGGUAAAACACUAGCCUUCCUUGUGCCCAUGCUAGAAAUCCUGCAGAGACGGCACAAG .((((((.....)))))).....(((((((.....((((((....))))))....))))(((((......))))))))...((((((((..(((.((.....))))).....)))))))) (-35.54 = -36.42 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:32 2006