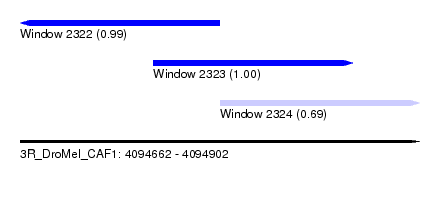
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,094,662 – 4,094,902 |
| Length | 240 |
| Max. P | 0.998560 |

| Location | 4,094,662 – 4,094,782 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -41.68 |
| Energy contribution | -41.76 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4094662 120 - 27905053 ACCCGCCACCGUUAUUGGAAAGGUAAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAGGCCCUGCCGCCUCUUUUGCCCAAACGCACAGUGCUAGGAAUCCACGGAAA ..(((..(((((((((.....)))))))))...........((((.(((((((((.....((((((.(((((......)))))....)))))).))))))).))))))......)))... ( -46.70) >DroSec_CAF1 6580 120 - 1 ACCCACCACCGAUAUUGGAAAGGUAAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAGGUCCUGCCGCCUCUUUUGCCCAAACGCACAGUGCUAGGAAUCCACGGAAA ........(((.....(((((((........)))))))...((((.(((((((((.....((((((.(((((......)))))....)))))).))))))).))))))......)))... ( -42.50) >DroSim_CAF1 6617 120 - 1 ACCCGCCACCGAUAGUGGAAAGGUAAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAGGCCCUGCCGCCUCUUUUGCCCAAACGCACAGUGCUAGGAAUCCACGGAAA ..(((((((.....))))...((...(((.........)))((((.(((((((((.....((((((.(((((......)))))....)))))).))))))).))))))...)).)))... ( -45.10) >DroEre_CAF1 6624 120 - 1 ACCCGAAACCGAUAUUGGUAAGGUGAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAAGCCCUGCCGCCUCUUCUGCCCAAACGCACAGUGCUAGGAAUCCACGGAAA ..(((..((((....)))).(((((..((.......)).)))))((((.((((((.....((((((.((((...........)))).)))))).))))))))))..........)))... ( -38.10) >DroYak_CAF1 6581 120 - 1 CCCCGCCACCGAUGUUGGAAAGGUGAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAGGCCCUGCCGCCUCUUCUGCCCAAACGCACAGUGCUGGGAAUCCACGGAAA ..(((.((((...........)))).(((.....(((((.....((((.((((((.....((((((.(((((......)))))....)))))).)))))))))).))))).))))))... ( -47.10) >consensus ACCCGCCACCGAUAUUGGAAAGGUAAUGGUCUUUUUCCCCACCUGCGCCUGUGUGGAGUAUUGGGCUGAGGCCCUGCCGCCUCUUUUGCCCAAACGCACAGUGCUAGGAAUCCACGGAAA ........(((.....(((((((........)))))))...((((.(((((((((.....((((((.(((((......)))))....)))))).))))))).))))))......)))... (-41.68 = -41.76 + 0.08)

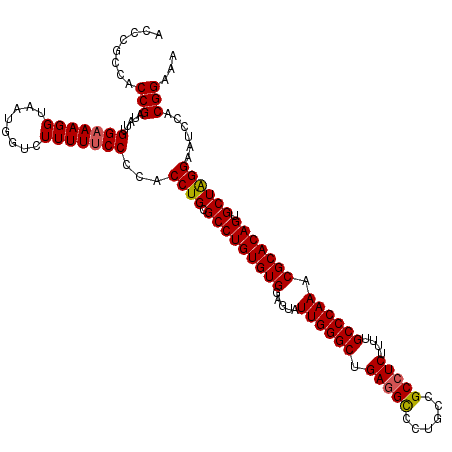

| Location | 4,094,742 – 4,094,862 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -40.46 |
| Energy contribution | -40.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4094742 120 + 27905053 GGGGAAAAAGACCAUUACCUUUCCAAUAACGGUGGCGGGUGAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGU .((........)).((((((..(((.......))).))))))........((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))) ( -43.60) >DroSec_CAF1 6660 120 + 1 GGGGAAAAAGACCAUUACCUUUCCAAUAUCGGUGGUGGGUGAACUGAGGAAUUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGU ...........((((((((...........))))))))............((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))) ( -42.60) >DroSim_CAF1 6697 120 + 1 GGGGAAAAAGACCAUUACCUUUCCACUAUCGGUGGCGGGUGAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGU .((........)).((((((..((((.....)))).))))))........((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))) ( -47.60) >DroEre_CAF1 6704 120 + 1 GGGGAAAAAGACCAUCACCUUACCAAUAUCGGUUUCGGGUGAACUCAGGAACUCCAGCAGGGCUACAAACUUCAGUUCUGGCUCGACAAUCCUGUAGAAAUUCUGCAGGCGGGCUGGCGU .((........)).((((((.(((......)))...))))))........((.(((((.((((((..(((....))).))))))..(...(((((((.....)))))))..)))))).)) ( -40.40) >DroYak_CAF1 6661 120 + 1 GGGGAAAAAGACCAUCACCUUUCCAACAUCGGUGGCGGGGGAACUAAGGAACUCCAGCAGGGCCACAAACUUGAGUUCUGGCUCGACAAUCCUGUAGAAAUUCUGCAGGCGGGCUGGCGU .((........)).((.(((..(((.......)))..)))))........((.(((((.((((((..(((....))).))))))..(...(((((((.....)))))))..)))))).)) ( -39.40) >consensus GGGGAAAAAGACCAUUACCUUUCCAAUAUCGGUGGCGGGUGAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGU .((........)).((((((..((......))....))))))........((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))) (-40.46 = -40.50 + 0.04)

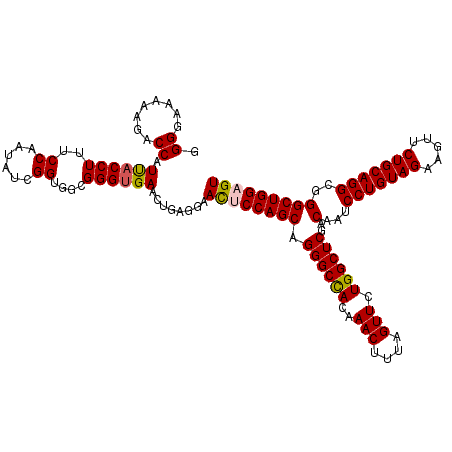
| Location | 4,094,782 – 4,094,902 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -36.92 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4094782 120 + 27905053 GAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGUGUUUACGGAUGCCUUCUCUUUGACUGAUACGAGAACAGGA ...(((..((((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))).))..)))...(((((((............))))..))). ( -42.40) >DroSec_CAF1 6700 120 + 1 GAACUGAGGAAUUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGUGUUUACGGAUGCCUUCUCUUUGACUGAUACGAGCACAGGA ...(((..((((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))).))..)))...(((.(((............)))...))). ( -38.10) >DroSim_CAF1 6737 120 + 1 GAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGUGUUUACGGAUGCCUUCUCUUUGACUGAUACGAGCACAGGA ...(((..((((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))).))..)))...(((.(((............)))...))). ( -40.40) >DroEre_CAF1 6744 120 + 1 GAACUCAGGAACUCCAGCAGGGCUACAAACUUCAGUUCUGGCUCGACAAUCCUGUAGAAAUUCUGCAGGCGGGCUGGCGUGUUCACGGAGGCCUUCUCCUUAACUGAAACGAGUACGGGA (.((((..((((.(((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))...))))..((((.....))))...........)))).).... ( -40.50) >DroYak_CAF1 6701 120 + 1 GAACUAAGGAACUCCAGCAGGGCCACAAACUUGAGUUCUGGCUCGACAAUCCUGUAGAAAUUCUGCAGGCGGGCUGGCGUGUUUACGGAGGCCUUCUCUUUAACUGAAACGAGUACAGGA ...((...((((.(((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))...))))....)).(((.((((((.....))).)))...))). ( -36.00) >consensus GAACUGAGGAACUCCAGCAGGGCCACAAACUUUAGUUCUGGCUCGACAAUCCUGUAGAAGUUCUGCAGGCGGGCUGGAGUGUUUACGGAUGCCUUCUCUUUGACUGAUACGAGCACAGGA ...(((.(((((((((((.((((((..(((....))).))))))..(...(((((((.....)))))))..))))))))).))).)))...(((.(((............)))...))). (-36.92 = -37.08 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:25 2006