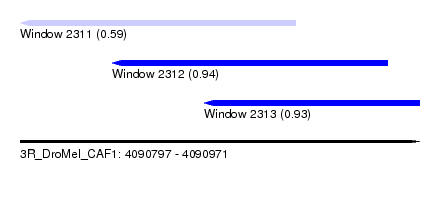
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,090,797 – 4,090,971 |
| Length | 174 |
| Max. P | 0.940088 |

| Location | 4,090,797 – 4,090,917 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -50.34 |
| Consensus MFE | -50.00 |
| Energy contribution | -49.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4090797 120 - 27905053 AAGUCGCCAACCGGCGACGUCAUUGCUGCCCGCAGAUCGCGGCGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGAACUGGGAUUGUCCAUUACGUGCGUCUGUUGUGCUUAGCGU .....((((((.(((((((((((..((((((((.....)))((((((((((....))))))))))....)))))..))))..((((....))))...))).)))).)))).))....... ( -52.00) >DroSec_CAF1 2711 120 - 1 AAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCCGCGGCAUCGCUCGGCGUAUUUGGCGGUGGUGAACUGGGAGUGUCCAUAAUGUGCGUCUGUUGUGCUUAGCGU ..((((((....)))))).....((((((((.((((((((.(((...))).))(((((.((.((....)).))))))))))))))).)).........(..((.....))..)..)))). ( -46.00) >DroSim_CAF1 2750 120 - 1 AAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCCGCGGCAUCGCUCGGCGUAUUUGGCGGUGGUGAACUGGGAGUGUCCAUAAUGUGCGUCUUUUGUGCUUAGCGU ..((((((....)))))).....((((((((.((((((((.(((...))).))(((((.((.((....)).))))))))))))))).)).........(..((.....))..)..)))). ( -46.00) >DroEre_CAF1 2766 120 - 1 AAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGACGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGAACUGGGAUUGUCCAUAACGUGCGUCUGUUGUGCUUAGCGU ..((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....))))))))))).(((((......(((((((......))))))))))))... ( -52.90) >DroYak_CAF1 2699 120 - 1 AAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGACGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGCGGUGAACUGGGAUUGUCCAUAACGUGUGUCUGUUAUGCUUAGCGU ..((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....))))))))))).(((((......(((((((......))))))))))))... ( -54.80) >consensus AAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGAACUGGGAUUGUCCAUAACGUGCGUCUGUUGUGCUUAGCGU ..((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....)))))))))))..((((....))))....(..((.....))..)....... (-50.00 = -49.92 + -0.08)

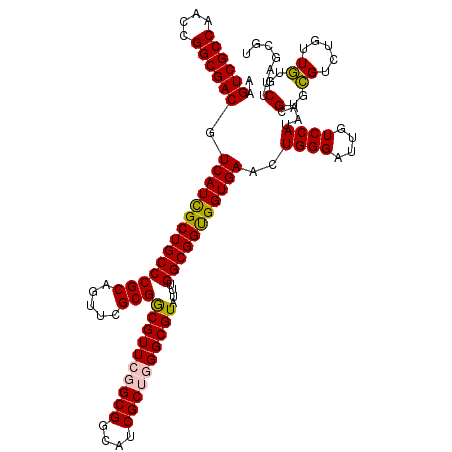
| Location | 4,090,837 – 4,090,957 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -50.90 |
| Consensus MFE | -51.42 |
| Energy contribution | -51.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4090837 120 - 27905053 CUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUUGCUGCCCGCAGAUCGCGGCGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((..((((((((.....)))((((((((((....))))))))))....)))))..)))) ( -52.60) >DroSec_CAF1 2751 120 - 1 CUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCCGCGGCAUCGCUCGGCGUAUUUGGCGGUGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((((((((((((.....)))(((((..(((....)))..)))))....))))))))))) ( -47.00) >DroSim_CAF1 2790 120 - 1 CUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCCGCGGCAUCGCUCGGCGUAUUUGGCGGUGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((((((((((((.....)))(((((..(((....)))..)))))....))))))))))) ( -47.00) >DroEre_CAF1 2806 120 - 1 CUGAUCUCCCAAGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGACGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....))))))))))) ( -53.00) >DroYak_CAF1 2739 120 - 1 CUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAAUGGCUCAAAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGACGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGCGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....))))))))))) ( -54.90) >consensus CUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGCGGCGUUCGGCGGCAUCGCUGGGCGUAUUUGGCGGUGGUGA ..(((((....)))))..(((.(((....)))..))).....((((((....)))))).((((((((((((((.....)))((((((((((....))))))))))....))))))))))) (-51.42 = -51.50 + 0.08)

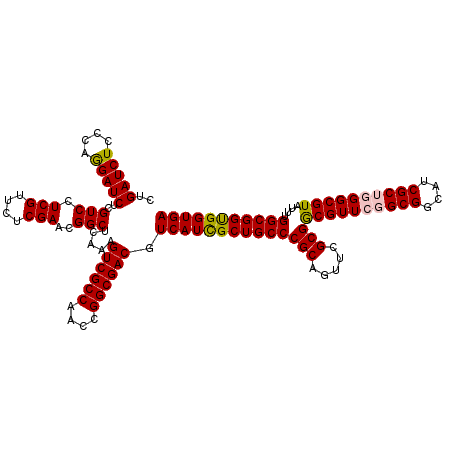
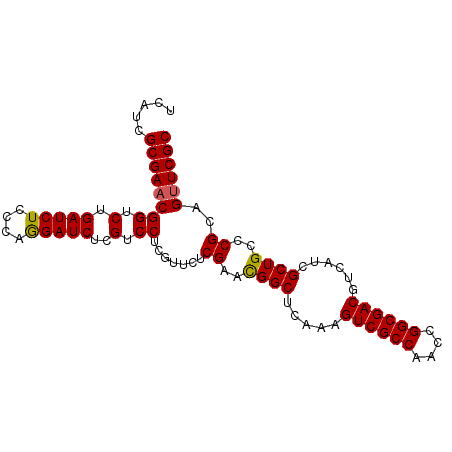
| Location | 4,090,877 – 4,090,971 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4090877 94 - 27905053 UCAUCGCGAACGGUCUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUUGCUGCCCGCAGAUCGC .....((((..((.(.(((((....)))))..).))....(((((..((((.....((((((....))))))......))))..)).))))))) ( -31.00) >DroSec_CAF1 2791 94 - 1 UCAUCGCGAACGGUCUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGC .....((((((((.(.(((((....)))))..).)).......((..((((.....((((((....))))))......))))..))..)))))) ( -31.80) >DroSim_CAF1 2830 94 - 1 UCAUCGCGAACGGUCUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGC .....((((((((.(.(((((....)))))..).)).......((..((((.....((((((....))))))......))))..))..)))))) ( -31.80) >DroEre_CAF1 2846 94 - 1 UCAUCGCGAACGGUCUGAUCUCCCAAGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGC .....((((((((.(.(((((....)))))..).)).......((..((((.....((((((....))))))......))))..))..)))))) ( -31.20) >DroYak_CAF1 2779 94 - 1 UCAUCGCGAACGGUCUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAAUGGCUCAAAGUCGCCCACCGGCGACGUCAUCGCUGCCCGCAGUUCGC .....((((((((.(.(((((....)))))..).)).((..(.(((.((((.....((((((....)))))))))))))..)..))..)))))) ( -29.90) >consensus UCAUCGCGAACGGUCUGAUCUCCCAGGAUCUCGUCCUCGUUCUCGAACGGCUCAAAGUCGCCAACCGGCGACGUCAUCGCUGCCCGCAGUUCGC .....((((((((.(.(((((....)))))..).)).......((..((((.....((((((....))))))......))))..))..)))))) (-30.72 = -30.60 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:14 2006