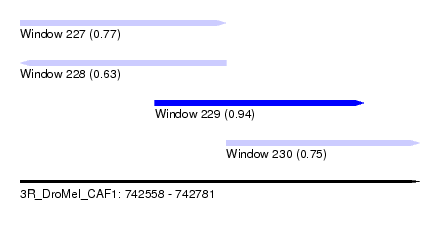
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 742,558 – 742,781 |
| Length | 223 |
| Max. P | 0.938346 |

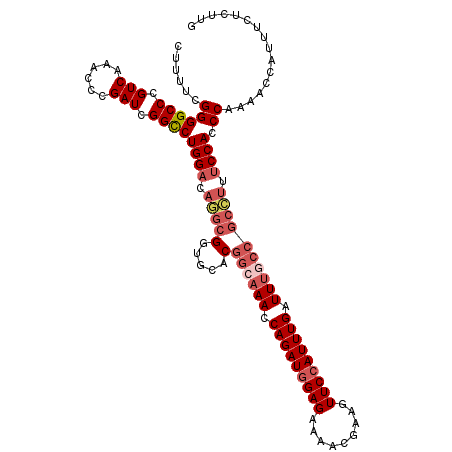
| Location | 742,558 – 742,673 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -27.86 |
| Energy contribution | -30.18 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 742558 115 + 27905053 CUUUUCGGGGCCCGUCAUACCCGAUCGGCCUGGACAAGCGGUGCACGGGAAACCAGAUAGAGAAACUGAACUUCCAUUUGAUUUGCCGCUUUUCCAGCCAAAACCAUUUCUCUUG ....(((((..........)))))..(((.((((.(((((((....((....))((((.(((.........))).)))).....))))))).)))))))................ ( -35.70) >DroSec_CAF1 9894 115 + 1 CUUUUCGGGGCCCGUCAAACCCGAUCGGUCUGGACAGGCGGUGCACGGCAAACCAGAUGGAGAAAACGAAGUUCCAUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUG ......((((((.(((......))).))).((((.(((((.....)((((((.(((((((((.........))))))))).)))))))))).)))))))................ ( -40.50) >DroSim_CAF1 9858 115 + 1 CUUUUCGGGGCCCGUCAAACCCGAUCGGUCUGGACAGGCGGUGCACGGCAAACCAGAUGGAGAAAACGAAGUUCCAUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUG ......((((((.(((......))).))).((((.(((((.....)((((((.(((((((((.........))))))))).)))))))))).)))))))................ ( -40.50) >DroEre_CAF1 10208 98 + 1 CUUUUCGGGGCCCGUCAAACCCGAUCGGCCUGG-----------------AAGCAGAUGGAGAAAACGAAGUUCCAUUUGAUUUGCCGCCUGUCCAGCCAGAACAAUUUCUCUUG ..((((.(((((.(((......))).))))).)-----------------)))(((((((((.........))))))))).........(((......))).............. ( -25.90) >consensus CUUUUCGGGGCCCGUCAAACCCGAUCGGCCUGGACAGGCGGUGCACGGCAAACCAGAUGGAGAAAACGAAGUUCCAUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUG ......((((((.(((......))).))))((((.(((((.....)((((((.(((((((((.........))))))))).)))))))))).)))).))................ (-27.86 = -30.18 + 2.31)

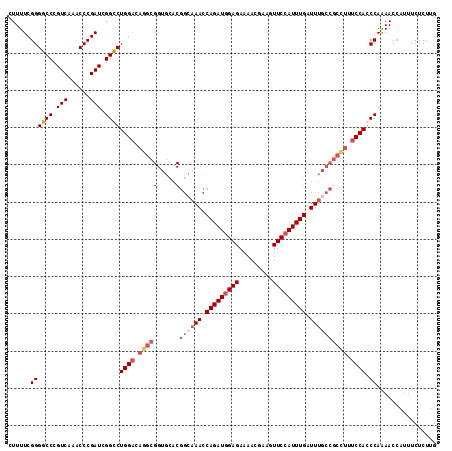
| Location | 742,558 – 742,673 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -28.91 |
| Energy contribution | -32.85 |
| Covariance contribution | 3.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 742558 115 - 27905053 CAAGAGAAAUGGUUUUGGCUGGAAAAGCGGCAAAUCAAAUGGAAGUUCAGUUUCUCUAUCUGGUUUCCCGUGCACCGCUUGUCCAGGCCGAUCGGGUAUGACGGGCCCCGAAAAG ............((((((..((....((((.((((((.(((((.(........)))))).)))))).))))...))(((((((...(((.....)))..))))))).)))))).. ( -33.10) >DroSec_CAF1 9894 115 - 1 CAAGAGAAAUGGUUUUGGGUGGAAAGGCGGCAAAUCAAAUGGAACUUCGUUUUCUCCAUCUGGUUUGCCGUGCACCGCCUGUCCAGACCGAUCGGGUUUGACGGGCCCCGAAAAG ............(((((((.((....(((((((((((.(((((...........))))).)))))))))))...))(((((((.(((((.....))))))))))))))))))).. ( -48.70) >DroSim_CAF1 9858 115 - 1 CAAGAGAAAUGGUUUUGGGUGGAAAGGCGGCAAAUCAAAUGGAACUUCGUUUUCUCCAUCUGGUUUGCCGUGCACCGCCUGUCCAGACCGAUCGGGUUUGACGGGCCCCGAAAAG ............(((((((.((....(((((((((((.(((((...........))))).)))))))))))...))(((((((.(((((.....))))))))))))))))))).. ( -48.70) >DroEre_CAF1 10208 98 - 1 CAAGAGAAAUUGUUCUGGCUGGACAGGCGGCAAAUCAAAUGGAACUUCGUUUUCUCCAUCUGCUU-----------------CCAGGCCGAUCGGGUUUGACGGGCCCCGAAAAG ....((((....))))(((((((..(((((........(((((...........)))))))))))-----------------))).)))..(((((..........))))).... ( -24.50) >consensus CAAGAGAAAUGGUUUUGGCUGGAAAGGCGGCAAAUCAAAUGGAACUUCGUUUUCUCCAUCUGGUUUGCCGUGCACCGCCUGUCCAGACCGAUCGGGUUUGACGGGCCCCGAAAAG ............(((((((.......(((((((((((.(((((...........))))).))))))))))).....(((((((.(((((.....))))))))))))))))))).. (-28.91 = -32.85 + 3.94)

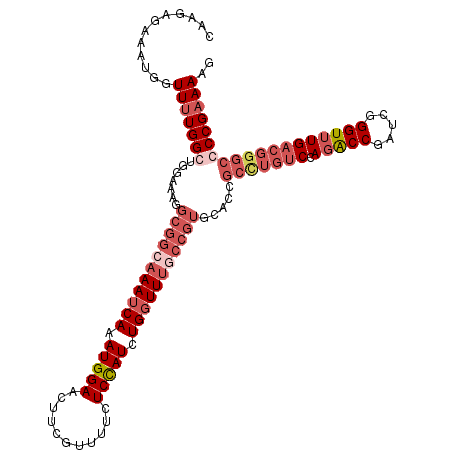
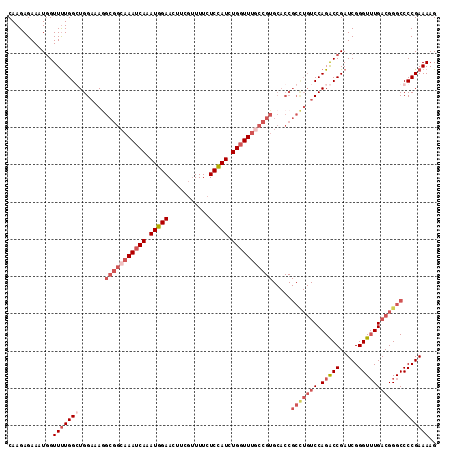
| Location | 742,633 – 742,750 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 742633 117 + 27905053 AUUUGAUUUGCCGCUUUUCCAGCCAAAACCAUUUCUCUUGUUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAUUUUCACAGGUGGCCCACUAAUUUCUUCCUACCAUUCCUC ......((((..(((.....))))))).......................(((((((((((.(((...............))).)))))))))))...................... ( -21.66) >DroSec_CAF1 9969 104 + 1 AUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUGCUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAUUUUCUUAGGUGGCCCACUAAUUUC-------------UC .........(((((((....(((((....(((...........))).....))))).((((.....))))..............)))))))...........-------------.. ( -20.80) >DroSim_CAF1 9933 104 + 1 AUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUGCUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAUUUUCUUAGGUGGCCCACUAAUUUC-------------UC .........(((((((....(((((....(((...........))).....))))).((((.....))))..............)))))))...........-------------.. ( -20.80) >DroEre_CAF1 10266 115 + 1 AUUUGAUUUGCCGCCUGUCCAGCCAGAACAAUUUCUCUUGUUUAUGCCCCGUGGGUCGCU-UUGAUAAGCUUAUUUAUUUUC-AAGGUGGCCCACUAAUUUUUUCCCACCACAUUUC ....((..((...........((..((((((......))))))..))...((((((((((-((((...............))-))))))))))))................))..)) ( -26.76) >consensus AUUUGAUUUGCCGCCUUUCCACCCAAAACCAUUUCUCUUGCUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAUUUUCUUAGGUGGCCCACUAAUUUC_____________UC ..................................................(((((((((((..((((((....)))))).....)))))))))))...................... (-18.20 = -18.45 + 0.25)


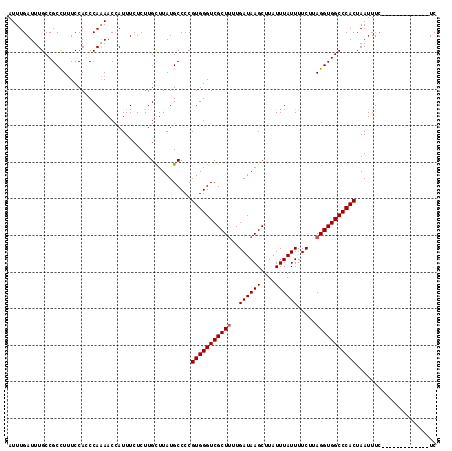
| Location | 742,673 – 742,781 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.65 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -12.13 |
| Energy contribution | -13.93 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 742673 108 + 27905053 UUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAU---UUUCACAGGUGGCCCACUAAUUUCUUCCUACCAUUCCUCAAUGUAGGUCUCUUUCGGU---UGUAGAUUGUGA (((((..((.(((((((((((.(((.............---..))).))))))))))).........(((((.(((....))))))))........)).---.)))))...... ( -28.46) >DroSec_CAF1 10009 95 + 1 CUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAU---UUUCUUAGGUGGCCCACUAAUUUC-------------UCAAUGUAGGUCUCUUUCGGU---UGUAGAUUGUCA ..........(((((((((((..((.............---..))..))))))))))).....((-------------(((((...((......)).))---)).)))...... ( -21.96) >DroSim_CAF1 9973 95 + 1 CUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAU---UUUCUUAGGUGGCCCACUAAUUUC-------------UCAAUGUAGGUCUCUUUCGGU---UGUAGAUUGUGA ..........(((((((((((..((.............---..))..))))))))))).....((-------------.((((.((.(.((.....)).---).)).)))).)) ( -23.36) >DroEre_CAF1 10306 109 + 1 UUUAUGCCCCGUGGGUCGCU-UUGAUAAGCUUAUUUAU---UUUC-AAGGUGGCCCACUAAUUUUUUCCCACCACAUUUCUAUUUAGGUCUCUUUUAUUAUGCUUAUAUUGUGA ..........((((((((((-((((.............---..))-))))))))))))..............((((........(((((............)))))...)))). ( -25.06) >DroYak_CAF1 10831 97 + 1 ------GUCAGUGGG-------UGAUAAGCUUAUAUAUAUAUUUA-CAGGUGGCCCAAUAAUUUCCUACUAUCAUUUCAAAAUUUAGACUCCUUUCGGU---CAUAGAUUGUGA ------.....((((-------(.((......(((....)))...-...)).)))))...................(((.((((((((((......)))---).)))))).))) ( -15.70) >consensus CUUAUGCCCCGUGGGUCGCUUUUGAUAAGCUUAUUUAU___UUUC_CAGGUGGCCCACUAAUUUC_U_C_A_CA____UCAAUGUAGGUCUCUUUCGGU___UGUAGAUUGUGA ..........(((((((((((..((..................))..)))))))))))........................................................ (-12.13 = -13.93 + 1.80)

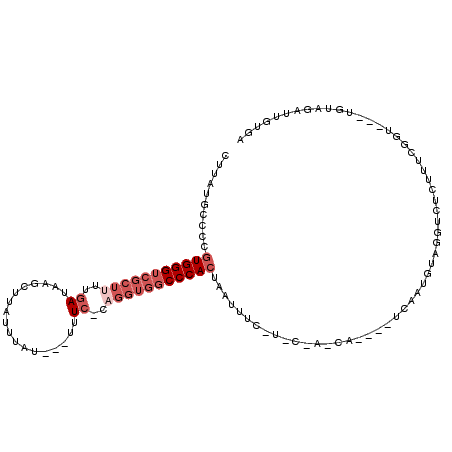
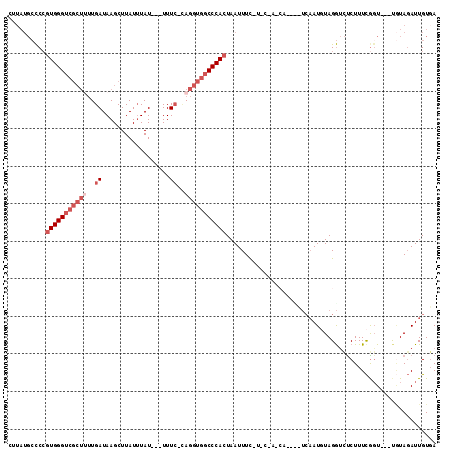
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:15 2006