
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,082,057 – 4,082,197 |
| Length | 140 |
| Max. P | 0.807922 |

| Location | 4,082,057 – 4,082,157 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -17.07 |
| Energy contribution | -18.73 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4082057 100 - 27905053 UAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACAC--AGGCAAAAAUUAUGG-------C----UAAAACUAUCUUAAAGU------- ........(((((((((((((.....)))))))(((((...((((...((((((...)))))).)))--)))))).....))))-------)----)................------- ( -27.90) >DroEre_CAF1 10830 120 - 1 UAAUGGCAAGCCAUUAUCUGCGAGUGGCACAUGUUGCCAAUUCUGGUAAUCGUUUUAGAUGAUACACUUGGACAAAAAUUAUGAACUAUGGCCCAGUAAAAGCAUAUUAAAAUACCUCCU ((((((....))))))..(((.(.(((..((((((((((....))))))).(((..((........))..)))..............)))..))).)....)))................ ( -22.20) >DroYak_CAF1 7717 101 - 1 UAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACACUUGGCCAAAAAUGACGGACUGUGGCCCAGUUAAA------------------- ....(((.((((..(((((((.....)))))))..(((((.((((...(((......)))..)))).)))))..........)).))...)))........------------------- ( -29.80) >consensus UAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACACUUGGACAAAAAUUAUGGACU_UGGCCCAGUAAAA__AU_UUAAA_U_______ .(((((....)))))((((((.....)))))).(((((((.((((...(((......)))..)))).)))).)))............................................. (-17.07 = -18.73 + 1.67)

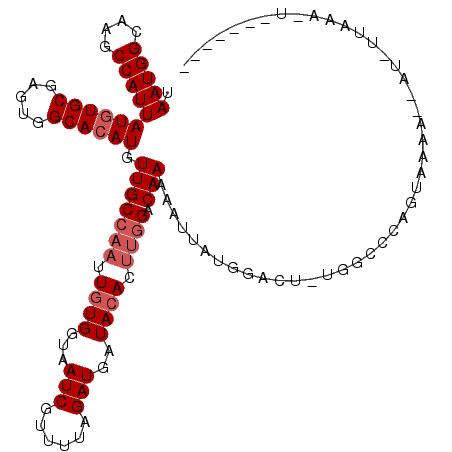
| Location | 4,082,079 – 4,082,197 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -31.27 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4082079 118 - 27905053 AUAUUCAUGGAGCAGCAUUUGAAACCGAUUCUAAGGCGGCUAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACAC--AGGCAAAAAUU ...........((..((((((((((.(((((((.((((((.(((((....)))))((((((.....)))))))))))).....))).))))))))))))))......--..))....... ( -34.10) >DroEre_CAF1 10870 120 - 1 AUAUUCAUGGAGCAGCAUUUGAAACCGAUUCUAAGGCGGCUAAUGGCAAGCCAUUAUCUGCGAGUGGCACAUGUUGCCAAUUCUGGUAAUCGUUUUAGAUGAUACACUUGGACAAAAAUU ...((((.(......((((((((((.(((((((..((((.((((((....)))))).))))((((((((.....)))).))))))).)))))))))))))).....).))))........ ( -35.70) >DroYak_CAF1 7738 120 - 1 ACAUUCAUGGAGCAGCAUUUGAAACCGAUUCUAAGGCGGCUAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACACUUGGCCAAAAAUG .((((..(((..(((((((((((((.(((((((.((((((.(((((....)))))((((((.....)))))))))))).....))).))))))))))))))......))).)))..)))) ( -36.80) >consensus AUAUUCAUGGAGCAGCAUUUGAAACCGAUUCUAAGGCGGCUAAUGGCAAGCCAUUAUGUGCGAGUGGCACAUGUUGCCAAUUGUGGUAAUCGUUUUAGAUGAUACACUUGGACAAAAAUU ...............((((((((((.(((((((.((((((.(((((....)))))((((((.....)))))))))))).....))).))))))))))))))................... (-31.27 = -31.60 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:11 2006